Control of dorsal mesoderm specification in Drosophila melanogaster by transcription factors networks.
2014/01/21
Abstract
During embryonic development, the action of a great number of transcription factors (TFs) and pathways enables the specification of a tissue and the cell differentiation. Here we focus on the Drosophila dorsal mesoderm which is well studied. At early stages, two pathways, the Decapentaplegic (Dpp) and Wingless (Wg) pathways as well as the Tinman TF characterize the dorsal mesoderm. They activate the expression of master regulators which control the development of different parts of the dorsal mesoderm. Indeed, combinations of master regulators TFs lead to the activation of a great number of enhancers that regulating other TFs and pathways proteins, enlarging the TFs network in cells. These TFs activate particular combinations of genes coding for proteins directly involved in cell differentiation.
Table of Contents
Introduction
How transcription factors (TFs) control embryogenesis is a longstanding and still poorly understood question. Mesoderm development is the best characterized model to answer it. The mesoderm is one of the three primary germ layers and is induced during the gastrulation. Mesoderm needs to be specified during early embryogenesis to subsequently differentiate into different subtypes that will give rise to various derived organs such as fat body, heart or muscles. An interesting model to study embryonic development is Drosophila melanogaster. Indeed, genetics and more recent molecular approaches allow describing quite in detail many actors involved in the different differentiation steps of mesoderm all along embryogenesis.
Drosophila mesoderm development proceeds in different phases. First of all, at stage 5, mesoderm is induced at the ventral pole of the embryo. At the next stage, it invaginates and extends along the anterior-posterior axis (Figure 1A). During these early stages, a few number of TFs expressed all over the mesoderm is induced. At stage 10, the Wingless (Wg) pathway is activated in stripes in the mesoderm, along the anterior-posterior axis. Then, at stage 11, stripes with the same identity migrate and merge together [1]. Wg pathway regulates specific TFs targets. So in the two domains, different TFs networks are established. These networks are controlled by master regulators TFs which activated more specialized genes inducing cell specific differentiation programs. TFs networks in the Wg- domain will give rise to the visceral mesoderm (splanchnopleure) whereas the Wg+ domain will form the cardiac and somatic mesoderm (somatopleure) (Figure 1B) [2]. The splanchnopleure differentiates into fat body, gonads and some somatic muscles [2,3], whereas somatopleure leads to the formation of different somatic muscles, of the heart (Figure 1C), which is the first embryonic tissue formed, and of heart associated tissues.
The TFs networks leading to cell differentiation has been first characterized by genetic analysis such as loss of function studies. These approaches have allowed to identified the master regulators of these networks. However redundant genes or genes inducing low phenotype had not been identified. New approaches of transcriptome and Chromatin Immuno-precipitation (ChIP) analysis allow to characterized all genes targeted by a master regulator TF and so belonging to the network that induced cell differentiation These studies have been performed on the dorsal mesoderm and wherein the focus will. Here we present how the dorsal mesoderm is specified over time, depending on signalling pathways and TFs networks.

Figure 1. Tinman expression is controlled by four enhancers that activate its expression in different regions over time. .
The genomic region with the four tin enhancers is represented on the left. The enhancers' names were given by Yin et al. [6] and are related to the domains in which they activate tin expression: Tin A = Anterior head element, Tin B = Broad mesodermal element, Tin C = Cardioblast element, Tin D = Dorsal mesoderm element. The TFs that inhibit the enhancers are represented in red and those that activate them in black. For three different stages, tin enhancers activation states and tin expression profiles are given. Tissues in which tin is expressed are represented on the embryos on the right. A Stage 6: The whole early mesoderm is delimited by a red line. Tin B is activated by Twist. Tin is expressed throughout the early mesoderm, except in the head mesoderm. Tin A is also activated by an unknown TF and tin is so expressed in the head too. B Stage 10: The dorsal mesoderm is delimited by a solid orange line and the ventral mesoderm by a dotted orange line. Tin B and Tin C are inhibited by Eve and Tin D is activated by Mad, Pnr and Tin itself. Tin D is repressed by Doc and Eve. Tin A is still active. So tin is expressed in the head and with fluctuations along the dorsal mesoderm. C Stage 16: The cardiac system is delimited by a yellow line. Tin B is still repressed by Eve. Tin D is inhibited by many TFs (Eve, Doc, Seven up (Svp), E(spl)-c) and Tin A is repressed by Buttonhead (btd). But Tin C is activated by unknown TFs. Tin expression is restricted to 4 cardiomyocytes and 2 pericardial cells per segment.
Early signals characterizing the dorsal mesoderm
At early stages of development, the patterning of the dorsal ventral axis in the Drosophila embryo is controlled by the Dorsal protein. Dorsal is present in a dorsal-ventral nuclear gradient. Indeed, at the ventral pole, Dorsal enters in the nucleus, whereas at the dorsal pole, Dorsal stays in the cytoplasm [4]. Dorsal acts as a morphogene: different target enhancers are activated depending on the Dorsal concentration along the dorsal-ventral axis. Enhancers with a low affinity for Dorsal are activated at the ventral pole by high concentrations of Dorsal. So, some genes are only activated at the ventral pole. This is the case for twist (twi) which is the inductor of the mesoderm [5]. The mesoderm is so induced in the ventral part of the embryo, at stage 5. Just after gastrulation, at stage 6, Twi activates Tinman (Tin) expression, another key regulator of the mesoderm (Figure 1A) [6]. This activation occurs after the Twi binding on one of its four enhancers and leads to a broad expression of tin throughout the mesoderm [6]. Tin is the vertebrate NK-2 homologue and binds target enhancers on the NK-2 motif with its homeodomain. Tin is regarded as a pioneer TF that induces chromatin remodeling which may possible the binding of other TFs [7]. Thereby, Tin gives the ability for the dorsal mesoderm cells to respond to the different specification signals. Then the Twi enhancer of tin is inhibited by the binding of the Even skipped (Eve) repressor and a second enhancer is activated (Figure 1B). This enhancer allows a dorsal mesoderm restricted expression [6]. So at stage 10, tin expression is restricted to the dorsal mesoderm.
Another signal characterizing the dorsal mesoderm is the decapentaplegic pathway (Dpp). Unlike twi, dpp gene is inhibited by high concentrations of Dorsal. As a consequence, dpp is more activated at the dorsal pole and so expressed in a reverse gradient of nuclear Dorsal along the dorsal-ventral axis [5]. Dpp acts as a morphogene too by activating genes depending on its concentration [8]. Dpp is only expressed in the ectoderm but diffuses within the underlying mesoderm and activates its own pathway into mesodermal cells. The activation occurs after the binding of the diffusible ligand Dpp on its receptor. This binding leads to the phosphorylation of the Mad protein, which recruits the Madea protein. The Mad/Madea complex penetrates into the nucleus and activates target genes. Dpp pathway is necessary for the specification of the dorsal mesoderm [9].
At stage 10, another important pathway is present in the dorsal mesoderm: the Wingless (Wg) pathway. The Wg pathway is activated by the binding of the Wg ligand on its receptor. Its effectors are the TF Pangolin (dTCF) and the two Sloppy Paired TFs: slp 1 and 2 [10]. The Wg ligand is secreted by the ectoderm and diffuses through five underlying cell layers within the mesoderm. But unlike Dpp, Wg ligand is expressed in stripes in the ectoderm [11]. As a consequence, the underlying dorsal mesoderm is striped by the Wg pathway in different patches along the anterior-posterior axis (Figure 2B) [10]. At stage 10, these Wg stripes are the only major difference observed within the dorsal mesoderm. But it leads to the specification between the visceral mesoderm, and the cardiac and somatic mesoderm.

Figure 2. Differential expression of master regulators in response to the Wg signal at stage 10. .
The occupancy and the activation state of enhancers of the master regulators eve, bap and pnr are given before and after the Wg induction, at stage 10. Embryos are represented in a sagittal plane, the dorsal mesoderm is delimited by a red line. A Before the Wg induction, Eve and Bap enhancers are repressed by an unknown TF whereas Pnr is activated by Tin and Dpp. B After Wg induction, the dorsal mesoderm is striped in Wg+ (purple) and Wg- (yellow) segments. The eve enhancer is activated by the binding of dTCF in Wg+ domains but is still repressed in Wg- domains. The bap enhancer is inactive in Wg+ domains because of the binding of the Slp repressor but bap is activated in Wg- domains. Pnr is still expressed in both domains.
Activation of master regulators involved in the dorsal mesoderm specification
The Wg pathway and its effectors induce changes in the activation state of many enhancers [12]. Some enhancers, inactive in the absence of Wg pathway are activated in Wg+ domains [10]. As a results the genes they control are expressed in stripes along the dorsal mesoderm. An example of these pair-rule genes is eve. During early stages Dpp effectors and Tin bind eve enhancer, but a repressor inhibits their activating action (Figure 2A). The binding of the Wg effector, dTCF, leads to an ousting of the repressor and an activation of the enhancer (figure 2B) [12]. Another master regulator of the somatopleure, the Dorsocross gene (Doc), is expressed in these Wg+ stripes. However Doc is only activated by Mad and dTCF, not by Tin [13]. These genes which are activated in the Wg+ domains, are master regulators of all somatic and cardiac mesoderm. Their activation is the first step of somatopleural specification.
In contrast, other enhancers are inhibited by the Wg pathway. Among them, some enhancers are already activated by Tin or Dpp effectors. Their activation is so restricted to the Wg- domain and the genes they control are expressed in negative stripes along the dorsal mesoderm. This is the case for bagpipe (bag) coding for a NK homeobox protein, like tin [14]. On the bap enhancer, the bindings of Tin and the Dpp effector Mad are sufficient for its activation. However the fixation of an effector of the Wg pathway, the repressor Slp, inhibits the bap enhancer (Figure 2B) [10,12]. The action of Slp may cause a chromatin remodeling or a recruitment of other repressors like Groucho, which represses the bap enhancer [12]. As a result, bap expression is restricted to the Wg- stripes. The same mechanism is observed on the biniou (bin) enhancer. Bin is a FoxF TF with a forkhead domain that regulates directly or indirectly 1164 genes, more than half of which negatively [15]. Biniou induces a positive feedback on the dpp and the bap enhancers [14]. The Wg- stripes will form the future visceral mesoderm and Bap and Bin are its master regulators.
Furthermore, the activation state of some enhancers may not be affected by the Wg pathway. Some of them are already activated by Tin and Mad. In this case they control genes which are expressed in the whole mesoderm, at stage 10. For example tin is still expressed at this stage throughout the mesoderm. This is also the case of pannier (pnr), expressed in the dorsal mesoderm since stage 8 (Figure 2). Pnr expression in the dorsal mesoderm had been first considered as a consequence of intermediate Dpp concentrations [16]. But a recent study has proved that the pnr enhancer is not activated by a Dpp threshold [8]. On its two enhancers, a direct Tin binding has been showed while Mad, despite having specific binding motifs on the enhancer, interacts with Tin protein to activate pnr transcription (Figure 2) [17]. Pnr codes for a GATA TF that binds Tin proteins in vivo and is so regarded as a Tin cofactor [8]. Pnr activates Dpp and Tin expression, in a positive feedback loop [18]. Tin and Pnr are master regulators of both splanchnopleure and somatopleure. However, pnr and tin are differentially regulated by the other master regulators and their targets along the anterior-posterior axis. For example, in the Wg+ patches, Doc and Eve inhibit directly tin expression [6]. On the other hand, Doc is an activator of pnr, a positive regulator of tin [13]. But ultimately, tin and pnr are more expressed in the Wg- than in the Wg+ cells (Figure 1B) [8].
The Wg pathway induces so a particular regulation of gene expression from stage 10. The expression of different TFs in the two domains of the dorsal mesoderm is the first step of specification. In the Wg+ stripes, Doc, Eve and low concentrations of Tin and Pnr control the differentiation program in somatic and cardiac muscles whereas in the Wg- stripes, Bin, Bap and high concentrations of Tin and Pnr induce the visceral mesoderm specification. Furthermore, pair-rule genes, expressed in stripes, are mutually exclusive, allowing a clear molecular distinction between the different bands [13].
Downstream networks inducing dorsal mesoderm specification
At stage 11, the stripes of same identity migrate and merge together [1]. But the morphological cell differentiation and organs formation occur later. The first larval differentiated organ is the heart and it takes shape only at stage 16. So between the stages 11 and 16, splanchnopleural and somatopleural master regulator TFs induce the activation of a large number of target genes leading to cell differentiation and organ formation.
The first identified target genes of early master regulators had been characterized one by one thanks to loss of function studies and only genes with high phenotype were studied. Genes with a redundant function or specialized genes had not been taken into account. This approach had limited the view of downstream networks [19,20]. Recent progress in ChIP and transcriptome analysis techniques allows the identification of a great number of target enhancers [15,21]. The target genes activated by these enhancers are then identified thanks to a computer program. Let us take the case of Tin: Tin is the first inductor of the dorsal mesoderm. In addition, Tin is a pioneer TF which recruits other TFs by fixating on enhancers. A technique of ChIP on chip allows to localize the different regions bound by Tin and the cofactors recruited. So ChIP on chip with Tin provides the different enhancers involved in the dorsal mesoderm specification and differentiation [21]. Tin regulated more than 480 enhancers involved in the regulation of approximately 260 genes [22].
In the Wg+ domain different enhancers are activated with particular combinations of TFs: enhancers only activated by Tin; enhancers activated by Tin, Doc and Pnr; enhancers activated by Tin, dTCF and Mad and enhancers activated by these five master regulators (Figure 3) [23]. These enhancers lead to the expression of specific somatopleural genes. Moreover, in the Wg+ domain, enhancers of splanchnopleural genes are occupied by these TFs but they are not active [24]. The binding of the Slp repressor inhibits them (Figure 3). On the other hand, in the Wg- domain, the bindings of the Pnr, Tin, Bin and Bap TFs allow the activation of enhancers of splanchnopleural genes (Figure 3). Conversely, in this domain, enhancers of somatopleural genes are inactive because of a lack of Eve, Doc or dTCF.
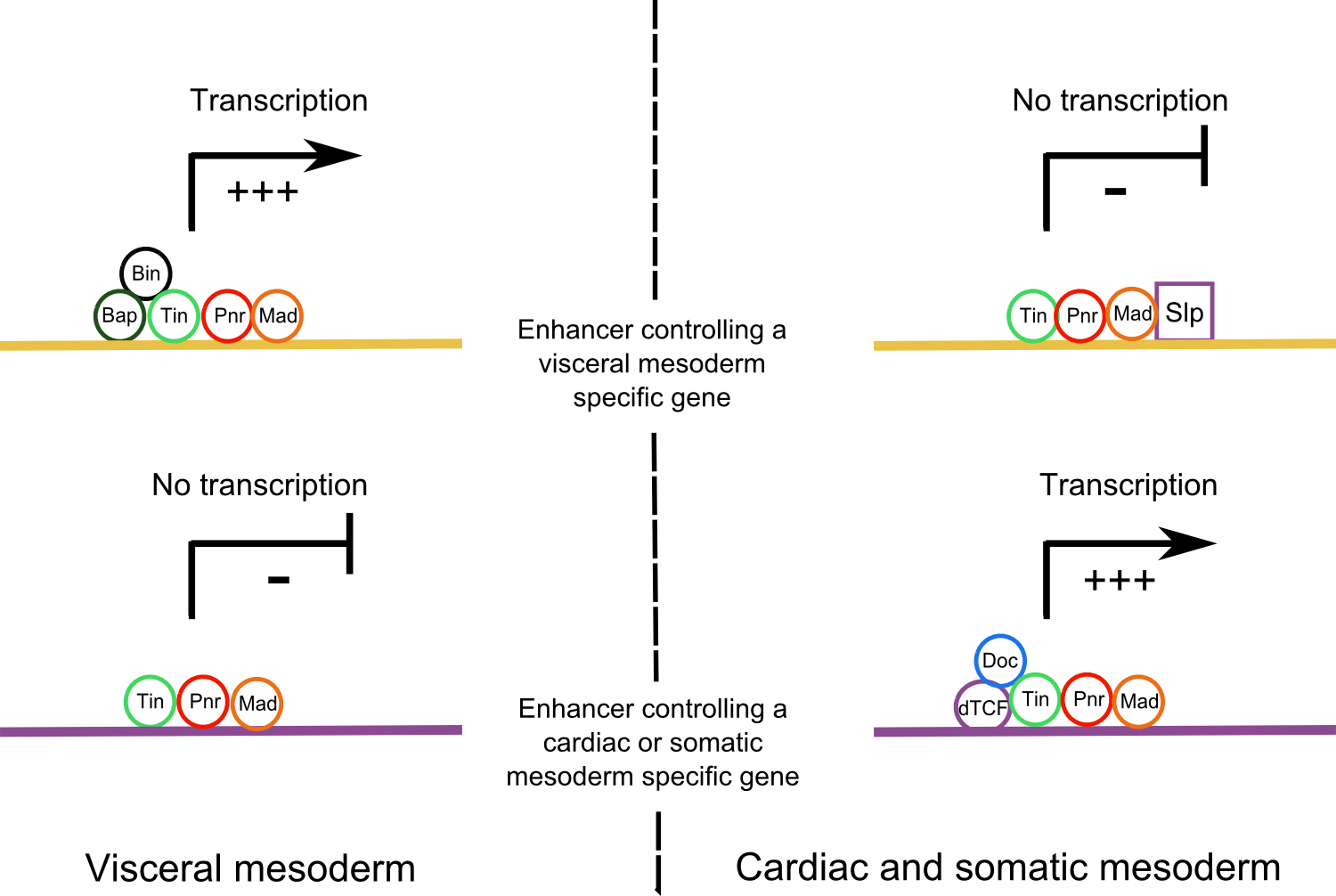
Figure 3. Activation of different downstream enhancers in Wg+ and Wg- domains. .
In the visceral muscles, enhancers that regulate a visceral specific gene are activated by a combination of different master regulators whereas in the cardiac and somatic mesoderm, the binding of the Slp repressor inhibits their expression. In opposite, enhancers that activate cardiac or somatic specific genes are activated by a combination of TFs in the cardiac and somatic mesoderm whereas they are not active in the visceral mesoderm because of a lack of necessary TFs.
In general, two different models are proposed to explain a TF binding on an enhancer: the enhanceosome model, in which the binding is highly cooperative and all proteins are fixed to one another; and the billboard model, in which the bindings of TFs are independent and the binding motifs order (grammar) on the enhancer is not very important [24]. Here bindings on enhancers are a mix of these two models. Tin is the first TF that binds on DNA. Then the other TFs are recruited cooperatively on the enhancer but the grammar of their bindings is not important [23]. The binding of a TF does not lead necessarily to the enhancer activation. Only a good combination of TFs on the enhancer can allow its activation [21,23].
Lots of genes are activated in the downstream networks. They code for proteins with manifold functions. These proteins could be classified into three categories: proteins involved in pathways, TFs controlling the differentiation of a cell type, and proteins which directly induce cell differentiation [14]. Indeed Tin regulates general pathways, the JAK/STAT and the Eya pathways, both involved in somatic muscles formation [22]. Transcription factors, regulating more specific differentiation processes in the dorsal mesoderm, such as Hand, Pointed or Mef2 are also activated [25–27]. These TFs are involved in positive or negative feedbacks, and feedforwards, which contribute to the robustness and the refinement of the processes [22]. Finally genes that code for proteins directly involved in the morphogenesis are activated in these networks too. For example a cytoskeleton protein, the β-3 Tubulin, has its expression directly activated by Pnr and Bin [14] and likewise the enhancer of katp, a gene coding for an ATP-sensitive potassium channel involved in cardiomyocytes development, is directly bound by Tin and Pnr [28]. Among the Tin and Pnr targets, we find genes coding for proteins involved in the actin cytoskeleton organization such as scr68B [23,29]. The timing of the enhancer binding by master regulators and downstream TFs is related to the function of the gene. General genes are activated during all the specification and differentiation steps in many cells whereas specific genes are expressed during late stages of differentiation in a low number of cells [15].
Unexpectedly, some early master regulators are recycled and play a role in the differentiation of specific tissues. For example, at stage 16, tin is only expressed in a few cardiomyocytes and pericardial cells per segment (Figure 1C) [13,30]. Tin expression is restricted by the action of different actors, like the TF Lame Duck, a target of the Dpp pathway [31], and later by Seven up and Doc, in the Tin- cardiomyocytes [30]. Interestingly a simple auto-restriction of tin expression is observed via the JAK/STAT signaling pathway. Tin activates the JAK/STAT pathway via Stat92E, which activates e(spl)-c genes that inhibites tin expression, inducing its restriction in some cardiomycytes. Twi expression is restricted by this same pathway at late stages too [32]. The tin restricted expression in four cardiomyocytes and a few number of pericardial cells per segment is permitted by the activation of another enhancer, that controls its late expression (Figure 2C). This late tin enhancer is regulated by Wg [6]. The same mechanism is observed for mef2. As in the vertebrates, mef2 controls the differentiation in all Drosophila larval muscles [33]. At early stages mef2 is expressed in the whole mesoderm, then in the dorsal mesoderm and finally in muscle precursors. Early roles of mef2 are not well known but at later stages, mef2 is the key regulator for the muscles differentiation. The mechanisms of regulation of mef2 expression are similar to the tin ones. First, after gastrulation, Twi binds a mef2 enhancer in the whole mesoderm. Then, the binding of Mad and Tin on a second enhancer, induce mef2 expression in the dorsal mesoderm. Later, in the different muscle precursors, mef2 expression is induced by specific combinations of TFs on different enhancers [25]. Eve and Doc are recycled too and play a role in the differentiation of a specific subset of cardiomyocytes. However, other early master regulators are not recycled. Bap expression stops just after dorsal mesoderm specification, whereas bin expression is maintained and not restricted in the whole visceral mesoderm, until stage 16 [15].
Combinations of late and early TFs lead to a subdivision within the splanchnopleure and somatopleure into different muscle and cardiac precursors. Precursor formation is the first step of differentiation in dorsal mesoderm. These precursors will induce the muscular cell proliferation and fusion, to build larval muscles.
Conclusion
The dorsal mesoderm specification is a well characterized model to understand the complexity of TFs networks leading to cell differentiation. During early stages of its development, only a low number of actors are involved and they are broadly expressed. At stage 10, the dorsal mesoderm is striped by alternating Wg+ and Wg- domains. The presence or the absence of the Wg pathway and so of their effectors Slp and dTCF is the only difference within the dorsal mesoderm. This little difference is responsible of an induction of different master regulators in the two domains. Then it is amplified thanks to the differential activation by these master regulators of a large number of enhancers. These enhancers control different types of genes: genes coding for pathway proteins, for TFs or for proteins directly involved in differentiation. These target genes are more specialized and their expressions are less broad. Furthermore, some master regulators are recycled and control the differentiation of a particular subset of cells. Our new knowledge on networks leading to the cell differentiation show that they are more complex than what was previously established. This is the result of a change in the techniques used for network characterization.
Indeed, during the 1990s and 2000s, studies of loss of function had enabled the identification of many master regulator genes. Then cis-regulatory networks had been established between them. But recent ChIP and transcriptomic studies show that their targets are very numerous. Feedforward and feedback loops make the prediction of their expression very complicated over time and space. Today, the biggest genes networks take into consideration 60 genes and all interactions are not well characterized [20]. In fact, during the differentiation stages, thousands of genes and hundreds of TFs interact within these networks. Current technologies make possible to study these networks at a cellular level, rather than at a tissue level [34,35]. Nucleus purification provides the epigenetic and transcriptomic landscapes for a particular cell [34]. One of the future challenges is to correctly characterize the trans-regulation of the different actors of these networks, and perhaps rebuild them numerically to predict gene expression in different cells.
Acknowledgments
I thank N. Fetter, L. Bloyet and Dr. S. Merabet for their help and their advice concerning this review.
References and recommended reading
Papers of particular interest have been highlighted as:
● of special interest
●● of outstanding interest
1. Azpiazu N, Frasch M: tinman and bagpipe: two homeo box genes that determine cell fates in the dorsal mesoderm of Drosophila. Genes Dev. 1993, 7:1325–1340. [PubMed]
2. Lawrence PA, Johnston P: Observations on cell lineage of internal organs of Drosophila. J. Embryol. Exp. Morphol. 1986, 91:251–266. [PubMed]
3. Clark IBN, Boyd J, Hamilton G, Finnegan DJ, Jarman AP: D-six4 plays a key role in patterning cell identities deriving from the Drosophila mesoderm. Dev. Biol. 2006, 294:220–231. [Full Paper][PubMed]
4. Kanodia JS, Rikhy R, Kim Y, Lund VK, DeLotto R, Lippincott-Schwartz J, Shvartsman SY: Dynamics of the Dorsal morphogen gradient. Proc. Natl. Acad. Sci. 2009, 106:21707–21712. [Full Paper][PubMed]
5. Sandmann T, Girardot C, Brehme M, Tongprasit W, Stolc V, Furlong EEM: A core transcriptional network for early mesoderm development in Drosophila melanogaster. Genes Dev. 2007, 21:436–449. [Full Paper][PubMed]
6. Yin Z, Xu XL, Frasch M: Regulation of the twist target gene tinman by modular cis-regulatory elements during early mesoderm development. Dev. Camb. Engl. 1997, 124:4971–4982. [PubMed]
7. Busser BW, Gisselbrecht SS, Shokri L, Tansey TR, Gamble CE, Bulyk ML, Michelson AM: Contribution of distinct homeodomain DNA binding specificities to Drosophila embryonic mesodermal cell-specific gene expression programs. PloS One 2013, 8:e69385. [Full Paper][PubMed]
8. Liang H-L, Xu M, Chuang Y-C, Rushlow C: Response to the BMP gradient requires highly combinatorial inputs from multiple patterning systems in the Drosophila embryo. Dev. Camb. Engl. 2012, 139:1956–1964. [Full Paper][PubMed]
9. Johnson AN, Burnett LA, Sellin J, Paululat A, Newfeld SJ: Defective decapentaplegic signaling results in heart overgrowth and reduced cardiac output in Drosophila. Genetics 2007, 176:1609–1624. [Full Paper][PubMed]
10. Lee et Frasch: Wingless effects mesoderm patterning and ectoderm segmentation events via induction of its downstream target sloppy paired. [date unknown], [no volume].
11. Riechmann V, Irion U, Wilson R, Grosskortenhaus R, Leptin M: Control of cell fates and segmentation in the Drosophila mesoderm. Dev. Camb. Engl. 1997, 124:2915–2922. [PubMed]
12. Lee H-H, Frasch M: Nuclear integration of positive Dpp signals, antagonistic Wg inputs and mesodermal competence factors during Drosophila visceral mesoderm induction. Dev. Camb. Engl. 2005, 132:1429–1442. [Full Paper][PubMed]
13. Reim I, Frasch M: The Dorsocross T-box genes are key components of the regulatory network controlling early cardiogenesis in Drosophila. Dev. Camb. Engl. 2005, 132:4911–4925. [Full Paper][PubMed]
14. Zaffran S, Küchler A, Lee HH, Frasch M: biniou (FoxF), a central component in a regulatory network controlling visceral mesoderm development and midgut morphogenesis in Drosophila. Genes Dev. 2001, 15:2900–2915. [Full Paper][PubMed]
15. Jakobsen JS, Braun M, Astorga J, Gustafson EH, Sandmann T, Karzynski M, Carlsson P, Furlong EEM: Temporal ChIP-on-chip reveals Biniou as a universal regulator of the visceral muscle transcriptional network. Genes Dev. 2007, 21:2448–2460. [Full Paper][PubMed]
16. Jazwinska A, Rushlow C, Roth S: The role of brinker in mediating the graded response to Dpp in early Drosophila embryos. Development 1999, 126:3323–3334. [PubMed]
17. Gajewski K, Zhang Q, Choi CY, Fossett N, Dang A, Kim YH, Kim Y, Schulz RA: Pannier is a transcriptional target and partner of Tinman during Drosophila cardiogenesis. Dev. Biol. 2001, 233:425–436. [Full Paper][PubMed]
18. Klinedinst SL, Bodmer R: Gata factor Pannier is required to establish competence for heart progenitor formation. Dev. Camb. Engl. 2003, 130:3027–3038. [PubMed]
19. Levine M, Davidson EH: Gene regulatory networks for development. Proc. Natl. Acad. Sci. U. S. A. 2005, 102:4936–4942. [Full Paper][PubMed][PubMed Central]
20. Bonn S, Furlong EEM: cis-Regulatory networks during development: a view of Drosophila. Curr. Opin. Genet. Dev. 2008, 18:513–520. [Full Paper][PubMed]
21. Jin H, Stojnic R, Adryan B, Ozdemir A, Stathopoulos A, Frasch M: Genome-wide screens for in vivo Tinman binding sites identify cardiac enhancers with diverse functional architectures. PLoS Genet. 2013, 9:e1003195. [Full Paper][PubMed]
22. Liu Y-H, Jakobsen JS, Valentin G, Amarantos I, Gilmour DT, Furlong EEM: A systematic analysis of Tinman function reveals Eya and JAK-STAT signaling as essential regulators of muscle development. Dev. Cell 2009, 16:280–291. [Full Paper][PubMed]
23. Junion G, Spivakov M, Girardot C, Braun M, Gustafson EH, Birney E, Furlong EEM: A transcription factor collective defines cardiac cell fate and reflects lineage history. Cell 2012, 148:473–486. [Full Paper][PubMed]
24. Zinzen RP, Girardot C, Gagneur J, Braun M, Furlong EEM: Combinatorial binding predicts spatio-temporal cis-regulatory activity. Nature 2009, 462:65–70. [Full Paper][PubMed]
25. Nguyen HT, Xu X: Drosophila mef2 expression during mesoderm development is controlled by a complex array of cis-acting regulatory modules. Dev. Biol. 1998, 204:550–566. [Full Paper]
26. Popichenko D, Sellin J, Bartkuhn M, Paululat A: Hand is a direct target of the forkhead transcription factor Biniou during Drosophila visceral mesoderm differentiation. BMC Dev. Biol. 2007, 7:49. [Full Paper][PubMed]
27. Alvarez AD, Shi W, Wilson BA, Skeath JB: pannier and pointedP2 act sequentially to regulate Drosophila heart. Development 2003, 130:3015–3026. [Full Paper][PubMed]
28. Akasaka T, Klinedinst S, Ocorr K, Bustamante EL, Kim SK, Bodmer R: The ATP-sensitive potassium (KATP) channel-encoded dSUR gene is required for Drosophila heart function and is regulated by tinman. Proc. Natl. Acad. Sci. U. S. A. 2006, 103:11999–12004. [Full Paper][PubMed][PubMed Central]
29. Djagaeva I, Doronkin S, Beckendorf SK: Src64 is involved in fusome development and karyosome formation during Drosophila oogenesis. Dev. Biol. 2005, 284:143–156. [Full Paper][PubMed]
30. Zaffran S, Reim I, Qian L, Lo PC, Bodmer R, Frasch M: Cardioblast-intrinsic Tinman activity controls proper diversification and differentiation of myocardial cells in Drosophila. Dev. Camb. Engl. 2006, 133:4073–4083. [Full Paper][PubMed]
31. Sellin J, Drechsler M, Nguyen HT, Paululat A: Antagonistic function of Lmd and Zfh1 fine tunes cell fate decisions in the Twi and Tin positive mesoderm of Drosophila melanogaster. Dev. Biol. 2009, 326:444–455. [Full Paper][PubMed]
32. Johnson AN, Mokalled MH, Haden TN, Olson EN: JAK/Stat signaling regulates heart precursor diversification in Drosophila. Dev. Camb. Engl. 2011, 138:4627–4638. [Full Paper][PubMed]
33. Soler C, Han J, Taylor MV: The conserved transcription factor Mef2 has multiple roles in adult Drosophila musculature formation. Development 2012, 139:1270–1275. [Full Paper][PubMed]
34. Steiner FA, Talbert PB, Kasinathan S, Deal RB, Henikoff S: Cell-type-specific nuclei purification from whole animals for genome-wide expression and chromatin profiling. Genome Res. 2012, 22:766–777. [Full Paper][PubMed][PubMed Central]
35. Southall TD, Gold KS, Egger B, Davidson CM, Caygill EE, Marshall OJ, Brand AH: Cell-type-specific profiling of gene expression and chromatin binding without cell isolation: Assaying RNA Pol II occupancy in neural stem cells. Dev. Cell 2013, 26:101–112. [Full Paper][PubMed][PubMed Central]


