Molecular and epigenetic regulations of the quiescent state in the adult mammalian stem cells
2014/01/21
Abstract
Adult stem cells have an essential role to maintain cellular homeostasis. Therefore the transition between the different states of these cells are controlled. This study focuses on the maintenance of the quiescent state, finely regulated by several molecular pathways and epigenetic phenomena. These intrinsic and extrinsic molecular signals combine to modulate cell division depending on extracellular conditions. The quiescent stem cells is well initiated by extracellular signals from the niche cells and maintained by various intracellular and epigenetic events which interact. Major signaling pathways affecting cell cycle are altered in quiescent cells in order to maintain the G0 stage.
Table of Contents
- Introduction
- Molecular regulations of the stem cell quiescent state
- Epigenetic modifications regulate the quiescent state of stem cells
- Discussion
- Acknowledgments
- References and recommended reading
Introduction
In adult, the homeostasis is maintained by a balance between proliferation and differentiation of stem cells. Adult stem cells are undifferentiated and multipotent cells, they can give rise to several types of cell when they differentiate to replenish dying cells and regenerate damaged tissues. These cells self-renew to maintain a pool of undifferentiated stem cells.
Adult stem cells are localized throughout the body in a specific microenvironnement named stem cell niche. Although the organization of stem cell niches seem diverse and induce different molecular interactions in function of the tissues, their role is to maintain stem cells in a functional state [1]. Physical and molecular interactions between stem cells and cells of the niche are responsible of the stem cell fate and their division plane, which determine the identity of the daughter cell [2]••.
Adult stem cells are very different from embryonic stem cells. The precise origin of adult stem cells in mature tissues is still unknown, while embryonic stem cells (ESC) derived from the inner cell mass of the blastocyst during embryogenesis. ESC are pluripotent, so they can give rise to all cell types, they are also able to divide indefinitely unlike adult stem cells whose self-renewing is limited.
That is why adult stem cells have a quiescent state to maintain a pool of cells that do not divide continuously, but may, in case of necessity, once again enter into the cell cycle (Box 1) and self-renew. In mammals, the niches contain quiescent and active stem cells specifically located [1]. Stem cells pass through a quiescent state by extrinsic signals mostly from the niche. Once quiescence is established, intrinsic and epigenetic molecular signals maintain this state. Stem cells can exit the G0 stage by receiving extrinsic molecular signals to enter into the mitotic cycle and divide when it is necessary to reestablish tissue homeostasis.
This stage of quiescence allows the maintenance of adult stem cells as well as cellular homeostasis, essential roles to preserve the tissue functions of an individual. In this review, we will explore what are the molecular – cellular components such as proteins – and epigenetic – heritable changes in gene activity regardless of changes in the DNA sequence – mechanisms, that regulate this quiescent state.
Molecular regulations of the stem cell quiescent state
A number of molecular signals regulate and maintain the quiescent state of adult stem cells. These signals can be intrinsic to the cell or extrinsic, from the microenvironment of stem cells and their interactions with their niche. The extrinsic signals initiates the transition from stem cells in a quiescent state.
Extrinsic mechanisms regulating stem cell quiescence
Maintaining the balance between stem cell quiescence and self-renewal is possible by signals emanating from the stem cell niche (Figure 1). The niche of hematopoietic stem cells (HSC), the more
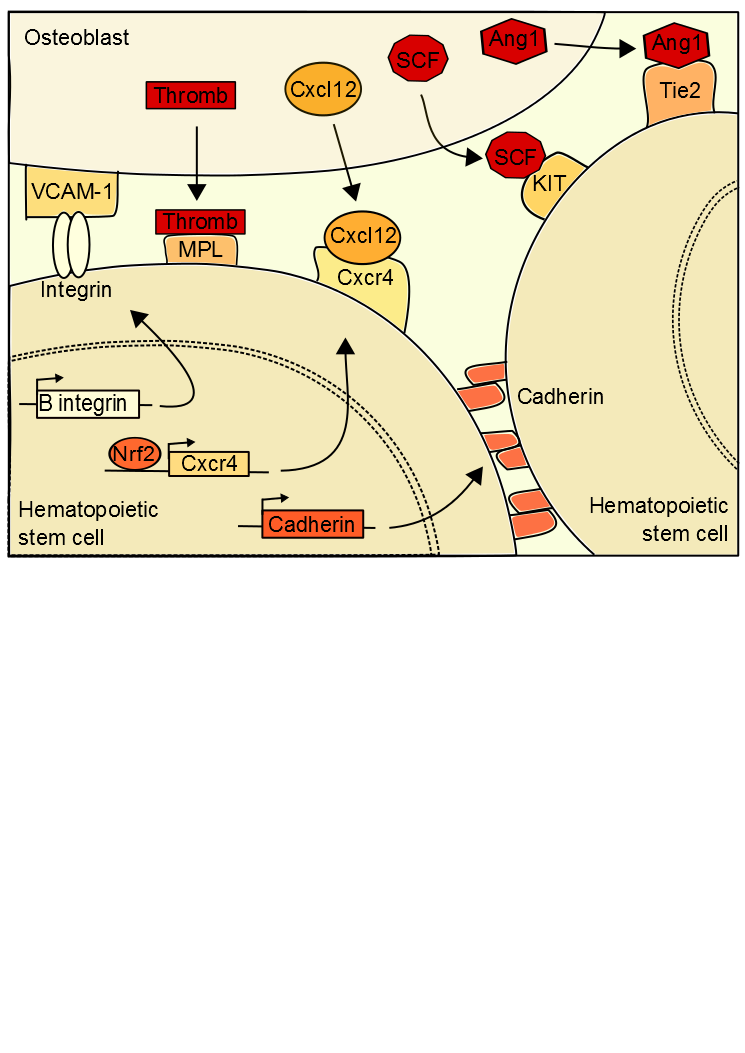
Figure 1. Extrinsic hematopoietic stem cell regulations. .
Quiescence is established in hematopoietic stem cells by interactions between stem cells and osteoblasts.
studied, is localized in the bone marrow (BM) [3]•. Adult hematopoietic stem cells are located in BM but they have an endothelial embryonic origin, results showed that mice HSC were produced in the aorta in the embryo, and then migrate into the BM. In adults, a microenvironment of osteoblasts, bone marrow stromal, endothelial cells and extracellular matrix surrounds the stem cells [4]. The state of HSC quiescence appears to depending on the location of stem cells in the niche. Interactions between cells induce HSC quiescent state, a retention of HSCs in their niche and a blocking of apoptosis. The hematopoietic microenvironment regulates HSC number and function for maintain the homeostasis. In case of homeostasis unbalance, expansions signals are forwarded to the stem cell so that it auto-renewed.
Signals from the niche are necessary to mediate the location of HSC.
CXCL-12 is a chemokine ligand produced by bone marrow and guiding HSCs to a specific niche location when bound to CXCR4 [5][6]••. In HSC, the activation of the expression of CXCR4 is dependent on Nrf2 (nuclear factor erythroid 2-related factor 2), a global regulator of oxidative stress response. Nrf2 binds to the CXCR4 gene promoter to activate its expression. Interaction between CXCL-12 and CXCR4 induced change in HSC niche location, which facilitates the HSC entry into quiescent state. This production of CXCL-12 is regulated by the sympathetic nervous system [7]••. A low level of CXCL-12 is correlated with an HSC output of the niche, inducing their differentiation. Therefore the expression of CXCL-12 is necessary to the establishment and maintenance of quiescent HSC. Moreover expression of CXCL-12 is mediated by genes of the molecular circadian clock, explaining the cyclical release of hematopoietic stem cells from the niche. The sympathetic nervous system secretes noradrenaline in the bone marrow that interacts with a osteoblast receptor inducing a down regulation of CXCL-12 expression [8]. Furthermore, integrin of the HSC binds to osteoblast VCAM-1 (Vascular cell adhesion protein 1), thereby preserving the location of the cell.
Quiescence state is possible by the presence of extrinsic signals.
Once the HSC at the right location, signals induce its quiescence. Ang-1 (Angiopoietin-1) binds to the stem cell receptor tyrosine kinase Tie2 which activates β‑integrin and N-cadherin expressions [9]. These proteins interact with the extracellular matrix and the niche cells inducing HSC retention. In addition the interaction between the thrombopotetin (thromb) and its MPL receptor (myeloproliferative leukemia virus oncogene) is also necessary to maintain the ideal number of hematopoietic quiescent stem cells, as the interaction between the osteoblast ligand SCF (Stem Cell Factor) and KIT receptor [10]. The osteopontin from the osteoblasts also inhibit the proliferation of the hematopoietic stem cells in the bone marrow niche. These various signals are anti-apoptosis, protect the cells from differentiation signals and arrest the cell-cycle.
Signals maintaining quiescence are in turn regulated.
The Wnt pathway regulates also the stem cells in the niche. By binding to Frizzled receptor, Wnt protein (wingless) initiates a signaling cascade inducing to stop the β-catenin degradation. The active signal of the β‑catenin induces the stem cell quiescence. Wnt signaling induces also an expression of p21, a CDKi, which inhibit the cell cycling. The expression of delta-like 1 homolog (Dlk1) from the osteoblasts inhibit the Wnt signaling and induce a proliferation of the HSC, cells out of their quiescent state [11, 12].
Role of some signals in the quiescent state regulation remains uncertain.
TGF-β and BMP signals from the osteoblasts seem maintain the quiescent state of the hematopoietic and neural stem cells by inhibiting the cell growth. However the publication of conflicting results indicates that other experiments may be necessary to understand the role of these molecules in the quiescence regulation.
Intrinsic mechanisms regulating stem cell quiescence
Although extrinsic signals induce quiescence of stem cells, the intrinsic molecular signals maintain this state. There is a strong link between the intrinsic and extrinsic regulation of quiescence. Extrinsic signals from the environment induce molecular cascades in stem cells which self-sustaining quiescence (Figure 2).
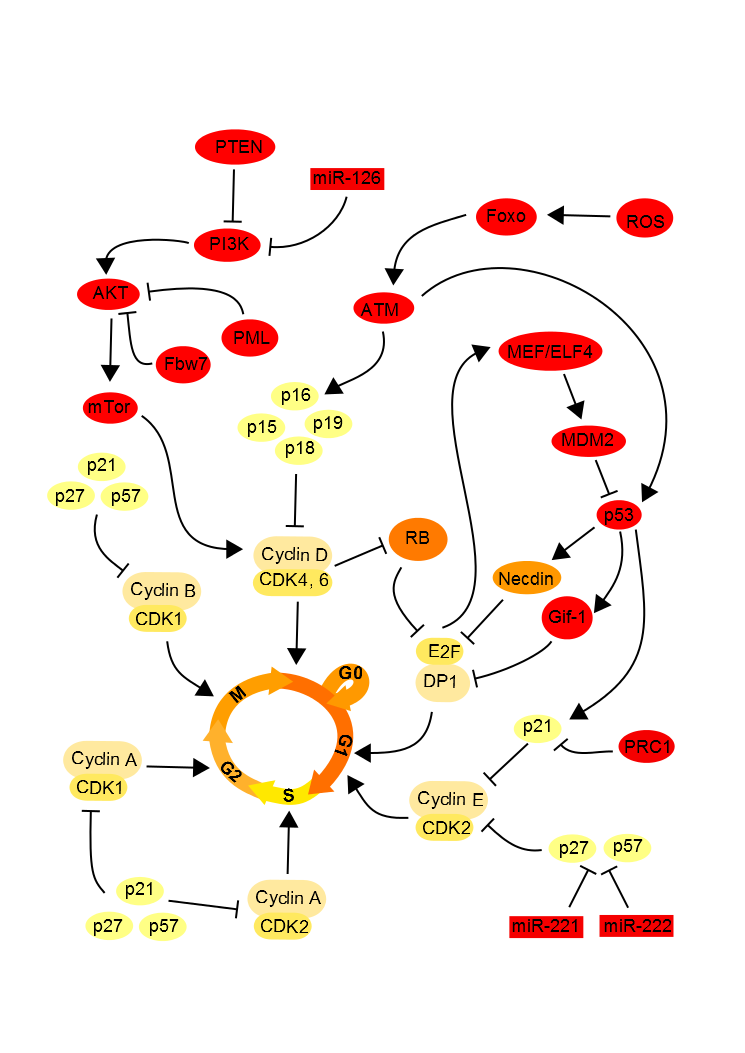
Figure 2. Cell cycle intrinsic regulation. .
The molecular signals are added to maintain the state of quiescence of hematopoietic stem cells.
Rb hypophosphorylation blocks the cell cycle.
The retinoblastoma (Rb) proteins family, known as tumor suppressors, possess a binding domain of many transcription factors, including E2F1. E2F1 is a transcription factor and this interaction with DP1 activates necessary genes to the cell to enter S phase in the cell cycle. It has been demonstrated that in the neural stem cells when the Rb protein is hypophosphorylated, it binds to E2F1 to repress and thus block the entry into S phase in the cell. In G1 phase, cyclin D and cyclin E activate the cyclin-dependent-kinases (CDK) CDK2 and CDK6 which will induce the phosphorylation of Rb. Once Rb phosphorylated, EF2 it releases which allows the cell cycle progress. In quiescent cells, Rb inhibits E2F, thereby blocking the cell cycle [13]••[14].
p53 up-regulates the quiescent state.
The transcription factor p53, known to block the cell cycle and induce apoptosis in case of DNA damage, promotes the quiescent state of neural and hematopoietic stem cells. P53 binds to promoters Gfi-1 (growth factor independent 1) and Necdin, a growth suppressor, to activate their expression. Thereby the expression level of Necdin is correlated with the rate of quiescent hematopoietic stem cells. Moreover, it has been shown in embryonic neural stem cells that Necdin can interact with the transcription factor E2F in the same way that the Rb family proteins, resulting in an inhibition of proliferation by blocking the cell cycle [15]. Similarly Gfi-1, which encodes a nuclear zinc finger protein that functions as a transcriptional repressor, limits the proliferation of these stem cells and maintain their integrity [16]. Interactions between Gfi-1, and several factors of the E2F family have also been identified, supposing that Gfi-1 regulates the cell cycle using this way. The p53 expression seems also modulated by the transcription factor MEF/ELF4 facilitating the entry of the hematopoietic stem cell into the cell cycle [13]••. E2F transcription factors enable MEF/ELF4 gene, allowing activation of MDM2, a repressor of p53. In quiescent cells, these factors E2F are inhibited, thereby blocking the cell cycle.
The CDKIs maintains the quiescent state.
The conservation of quiescent stem cells is also possible through inhibition of CDKs by CDKIs as p21, p27 and p57 [17]. P27 and p57 prevent the nuclear transport of the complex Hsp70-cyclin D1 necessary to advanced cell cycle and thus blocks the cell cycle in the hematopoietic stem cells. P21 is also necessary to keep the quiescent state of stem cells as it governs the entry into the cell cycle. In the neural stem cell, p15, p21, p16 and p27 suppress cell proliferation by inhibiting the phosphorylation of Rb, which can thus inhibit E2F.
Regulation of oxidative stress is necessary to maintain the quiescent state.
The intracellular level of reactive oxygen species (ROS) correlates with the proliferation of hematopoietic stem cells. Too high levels of ROS are toxic to the cell, but ROS is necessary to optimize the proliferation and differentiation. Thus, if the rate of ROS is too low, the cell enters a quiescent state. In the quiescent cell, Forkhead box O protein family (FOXOs) are activated in response to oxidative stress inducing an increase in expression of genes down-regulating the oxidative stress. ATM (ataxia telangiectasia mutated) and its target, p16 are downstream of FOXO and also repress the cell cycle [19, 20]. The inhibition by Rb to block the cell cycle is also dependent on the inhibition of oxidative stress by ATM. The AKT/PI3K, in link with a high level of ROS, induces the mTor pathway, responsible of cell proliferation [21]. So, for maintaining the quiescent state, the E3 ubiquitin ligase Fbw7 [22], PTEN [23] and PML [24] inhibit the mTor pathway in addition to regulating the ROS level.
Epigenetic modifications regulate the quiescent state of stem cells
The quiescent state of adult stem cells is also maintained by various epigenetic signals in complement of molecular signals (Box 2).
DNA methylation
DNA methylation of the neural stem cells is dynamic and active during the stage of quiescence, since the marks of DNA methylation are generally repression signals, it is assumed that the genes methylated are implicated in the differentiation. Inactivation of these genes induces maintenance of quiescent stem cells.
The pattern of imprinted gene expression, genes expressed in a parent-of-origin-specific manner, is largely retained in hematopoietic stem cells, as well as muscle and epidermal stem cells. Genes subject to imprints as p57, or Necdin have a role in quiescence maintaining.
Maternal imprinting at the H19–Igf2 locus maintains the quiescent state.
In the hematopoietic stem cells, the maternal H19 region of methylated DNA allows the production of miR-675 [25]. This miR-675 limits the expression of IGF1R (insulin-like growth factor 1 receptor), a factor in the mTor pathway that promotes proliferation. Therefore if miR-675 limits the proliferation by blocking IGF1R, quiescence is maintained.
PIAS1 maintains the quiescent state of hematopoietic stem cells.
PIAS1 (Protein inhibitor of activated STAT-1), a SUMO E3 ligase-protein inhibits DNMT3A, a DNA-methyltransferase, and STAT1, an activator of transcription. These inhibitions induce methylation and inactivation of GATA1 and of many other genes to keep the quiescent state of the hematopoietic stem cell [26,27].
According to current studies, CpG methylation marks seem especially differ between undifferentiated state and differential stem cells rather than on their quiescent or proliferation state [28]. However, it should be noticed that quiescent stem cells have a different expression profile for imprinted genes, therefore dependent gene methylation.
Small interfering RNAs
Many miRNAs regulate the quiescent stem cells by inhibiting molecules inducing cell differentiation or proliferation.
MiR-489 maintains the quiescent state by inactivation of Dek.
In the muscle stem cells, the rate of miRNA-489 expression is correlated with the state of the stem cells, a high miRNA expression indicates cell quiescence [29]. The target of this miRNA is Dek, an oncogene involved in chromatin organization to facilitate transcription and induce a cell proliferation and regulating the exit from quiescence. Consequently, by the inhibition of Dek, the miRNA-489 preserve the quiescent state.
MiR-126 controls the quiescent state by the regulation of the PI3K–AKT signaling pathway.
In HSC, miRNA-126 inhibits PI3K/AKT pathway and blocks mTor downstream PI3K. Since mTor pathway induces cell proliferation, block this signaling by miR-126 maintains quiescence [30].
MiR-31 is present in the quiescent stem cells.
In quiescent muscle cells, the Myogenic factor Myf5 which regulates muscle differentiation is transcribed but not translated. In fact, the Myf5 messenger RNA (mRNA) is sequestered and targeted by miR-31, which silences it. When the cell is activated, Myf5 mRNA is released in order to be transcribed and induce differentiation or myogenesis [31]. Therefore the miR-31 is transcribed in quiescent cells to inhibit cell differentiation.
MiR-221 and miR-222 prevent quiescence state.
The miR-221 and miR-222 target the p27 and p57 CDKIs to induce the transition from a quiescent cell in the S phase of the cell cycle [32]. These miRNAs inhibit cell cycle inhibitors, the cycle is active and the cells divide.
Histones modifications
Quiescent neural stem cells have a complex epigenetic pattern.
They have on the same gene stimulatory (H4ac) and inhibitory signals (H3Kme) [34]. The presence of two types of signals suggests the necessity of a fine and fast control. To exit the quiescent state it is necessary to remove some inhibitory marks so that chromatin becomes accessible to the transcriptional machinery. H3K4me2 and H3K27me3 epigenetic marks are higher in quiescent neural cells and reduction of these marks is correlated with cell differentiation. So, these epigenetic mark modifications are associated with genes to maintain the quiescent state of the cells. During the quiescent state, the genes required for cell differentiation are inhibited by chromatin compaction thanks to epigenetic histone marks.
H4K20me2 and H4K20me3 are associate with the quiescent state.
H4K20 methylation is the mark most significantly enriched in chromatin of quiescent stem cells. The methylation decrease at this histone site is correlated with cell proliferation. The methylation of H4K20 plays on the compaction of DNA, therefore more this lysine is methylated, more the DNA is compact and thus impossible to transcribe [34].
Polycomb group
The polycomb complex PRC1 is a marker of active state.
In the neural stem cells in culture, the polycomb repressive complex 1 (PRC1) component Bmi1 repress p21, the CDKI p16 and p19. However these CDKIs are required to maintain the quiescent state of stem cells. The regulation of Polycomb group therefore appears to induce in the neural stem cells an activation and differentiation of stem cells. In HSC the PRC1 inhibit the Ink4/Arf locus, which encodes cell cycle inhibitors p16 and p19. Since inhibitors cycle are inhibited, cell proliferation is induced. Bmi1 prevents their differentiation so the stem cells can self-renew and proliferate [35,36].
EZH2 influences the self-renewal and proliferation of the stem cells.
EZH2, a subunit of the Polycomb repressive complex 2, is not expressed in quiescent cells. However, the presence of EZH2 in the stem cells is correlated with the activation of this stem cells. So EZH2 appears required to reactivate the cell cycle [37,38]. EZH2 also plays an important role in the proliferation and differentiation but their functions are not yet clear, further studies would be necessary to better understand this regulatory mechanism.
The various polycomb complexes appear to be characteristic of active stem cells because they induce the inhibition of factors maintaining the quiescent stem cell state.
The epigenetic regulation is a combination of this different marks
By the example of epigenetic regulation inducing senescence of cancerous stem cells can be seen that all regulation phenomena are dependent [39]•. The inhibition of DNA methyltransferases (DNMT) by small interfering RNA in the active cells can induce a modification on histone marks and a decrease of methylation in CpG islands in the promoters of p16 and p21. Inhibition of DNMT also induces an increase in miRNA targeting polycomb complex, and therefore a reduction of complexes, including Bmi1 and EZH2, which methyl the H3K9 and H3K27 on the promoters of p16 and p21. There is therefore a reactivation of these CDKIs, which will block the cell cycle in G1 phase inducing cell senescence.
Although molecular actors appear identical between the phenomenon of senescence and quiescence, the cellular senescence regulatory varies because is not reversible unlike quiescence. Over a senescent cell is a damaged cell, while the cell quiescence is a stage that can be intermediate, wherein the cell is not dividing.
Discussion
The mechanisms establishing the quiescent stem cells are extracellular and dependents of the niche. Soon as the cell is in a state of quiescence, many molecular and epigenetic factors maintain this state. The cells appear to leave the quiescent state only by restimulation from the cells of the niche. The regulation of quiescence is accomplished in the microenvironment of stem cells but also by more distant signals from the nervous system to maintain cellular homeostasis.
Many diseases are associated with stem cell dysregulations and therefore results in a disequilibrium of tissue homeostasis. A modification of expression of factors that regulate the cell cycle induces changes in cell proliferation rate, jeopardizing cellular homeostasis. Many factors known to be tumor suppressors or oncogenes are involved in the regulation of the quiescent state, if these factors are not tightly regulated, an excessive cell proliferation may develop and cause cancer. It has been shown that problems of cell proliferation regulation may induce cancers such as leukemia if it is the hematopoietic stem cells that are affected [40], or melanoma for epithelial stem cells [41]. Control quiescence is one of the tracks in the development of new cancer treatments. Indeed, mutated stem cells entering into quiescence are not affected by current chemotherapy treatments and are a pool of cells able to create new tumors if they emerge from their quiescence [42].
Understanding the regulation of stem cells is also essential to succeed the implementation of cell-based therapies aimed at restoring a tissue or organ by grafting new cells. Moreover, the transformation of adult stem cells in induced pluripotent cells also requires a perfect understanding of the mechanisms regulating these cells.
Acknowledgments
I thank Dr Valerie Goubard for her help and advice concerning this review.
References and recommended reading
Papers of particular interest have been highlighted as:
● of special interest
●● of outstanding interest
1. Mitsiadis TA, Barrandon O, Rochat A, Barrandon Y, De Bari C: Stem cell niches in mammals. Exp. Cell Res. 2007, 313:3377–3385. [Full Paper][PubMed]
2. Li
L, Xie T: Stem cell niche:
structure and function.
Annu. Rev. Cell Dev.
Biol. 2005, 21:605–631.
[Full Paper][PubMed] ●● This is a full review on
the functions of different types of stem cell
niches.
3.
Wilson A, Trumpp A: Bone-marrow haematopoietic-stem-cell niches. Nat. Rev.
Immunol. 2006,
6:93–106. [Full Paper][PubMed].
● A specific review to hematopoietic stem cells that
are most often studied to understand the mechanisms of quiescence.
4. Calvi LM, Adams GB, Weibrecht KW, Weber JM, Olson DP, Knight MC, Martin RP, Schipani E, Divieti P, Bringhurst FR, et al.: Osteoblastic cells regulate the haematopoietic stem cell niche. Nature 2003, 425:841–846. [Full Paper]
5. Nie Y, Han Y-C, Zou Y-R: CXCR4 is required for the quiescence of primitive hematopoietic cells. J. Exp. Med. 2008, 205:777–783. [Full Paper][PubMed][PubMed Central]
6.
Tzeng Y-S, Li H, Kang Y-L, Chen W-C, Cheng W-C, Lai D-M:
Loss of Cxcl12/Sdf-1 in adult mice
decreases the quiescent state of hematopoietic stem/progenitor cells and alters
the pattern of hematopoietic regeneration after
myelosuppression.
Blood 2011, 117:429–439. [Full Paper][PubMed] ●● CXCL12 the signal is one of the most important
signals to the input of the quiescent stem cells.
7.
Katayama Y, Battista M, Kao W-M, Hidalgo A, Peired AJ, Thomas SA,
Frenette PS: Signals from the
sympathetic nervous system regulate hematopoietic stem cell egress from bone
marrow. Cell 2006,
124:407–421. [Full Paper][PubMed] ●● Article which makes the link between the nervous
system and the microenvironment of stem cells, ranging up to molecular
changes
8. Méndez-Ferrer S, Lucas D, Battista M, Frenette PS: Haematopoietic stem cell release is regulated by circadian oscillations. Nature 2008, 452:442–447. [Full Paper][PubMed]
9. Gomei Y, Nakamura Y, Yoshihara H, Hosokawa K, Iwasaki H, Suda T, Arai F: Functional differences between two Tie2 ligands, angiopoietin-1 and -2, in regulation of adult bone marrow hematopoietic stem cells. Exp. Hematol. 2010, 38:82–89. [Full Paper][PubMed]
10. Yoshihara H, Arai F, Hosokawa K, Hagiwara T, Takubo K, Nakamura Y, Gomei Y, Iwasaki H, Matsuoka S, Miyamoto K, et al.: Thrombopoietin/MPL signaling regulates hematopoietic stem cell quiescence and interaction with the osteoblastic niche. Cell Stem Cell 2007, 1:685–697. [Full Paper][PubMed]
11. Ferrón SR, Charalambous M, Radford E, McEwen K, Wildner H, Hind E, Morante-Redolat JM, Laborda J, Guillemot F, Bauer SR, et al.: Postnatal loss of Dlk1 imprinting in stem cells and niche astrocytes regulates neurogenesis. Nature 2011, 475:381–385. [Full Paper][PubMed][PubMed Central]
12. Fleming HE, Janzen V, Lo Celso C, Guo J, Leahy KM, Kronenberg HM, Scadden DT: Wnt signaling in the niche enforces hematopoietic stem cell quiescence and is necessary to preserve self-renewal in vivo. Cell Stem Cell 2008, 2:274–283. [Full Paper]
13.
Taura M, Suico MA, Fukuda R, Koga T, Shuto T, Sato T, Morino-Koga S,
Okada S, Kai H: MEF/ELF4
transactivation by E2F1 is inhibited by p53. Nucleic
Acids Res. 2011,
39:76–88. [Full Paper][PubMed] ●● Section that connects different tumor suppressor
factors, stem cell quiescence and regulation of the cell cycle
14. Weinberg RA: The retinoblastoma protein and cell cycle control. Cell 1995, 81:323–330. [PubMed]
15. Asai T, Liu Y, Di Giandomenico S, Bae N, Ndiaye-Lobry D, Deblasio A, Menendez S, Antipin Y, Reva B, Wevrick R, et al.: Necdin, a p53 target gene, regulates the quiescence and response to genotoxic stress of hematopoietic stem/progenitor cells. Blood 2012, 120:1601–1612. [Full Paper][PubMed]
16. Purizaca J, Contreras-Quiroz A, Dorantes-Acosta E, Vadillo E, Arriaga-Pizano L, Fuentes-Figueroa S, Villagomez-Barragán H, Flores-Guzmán P, Alvarado-Moreno A, Mayani H, et al.: Lymphoid progenitor cells from childhood acute lymphoblastic leukemia are functionally deficient and express high levels of the transcriptional repressor Gfi-1. Clin. Dev. Immunol. 2013, 2013:349067. [Full Paper][PubMed][PubMed Central]
17. Lee J, Hoi CSL, Lilja KC, White BS, Lee SE, Shalloway D, Tumbar T: Runx1 and p21 synergistically limit the extent of hair follicle stem cell quiescence in vivo. Proc. Natl. Acad. Sci. U. S. A. 2013, 110:4634–4639. [Full Paper][PubMed][PubMed Central]
18. Tothova Z, Kollipara R, Huntly BJ, Lee BH, Castrillon DH, Cullen DE, McDowell EP, Lazo-Kallanian S, Williams IR, Sears C, et al.: FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell 2007, 128:325–339. [Full Paper][PubMed]
19. Ito K, Hirao A, Arai F, Matsuoka S, Takubo K, Hamaguchi I, Nomiyama K, Hosokawa K, Sakurada K, Nakagata N, et al.: Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature 2004, 431:997–1002. [Full Paper][PubMed]
20. Furukawa-Hibi Y, Yoshida-Araki K, Ohta T, Ikeda K, Motoyama N: FOXO forkhead transcription factors induce G(2)-M checkpoint in response to oxidative stress. J. Biol. Chem. 2002, 277:26729–26732. [Full Paper][PubMed]
21. Juntilla MM, Patil VD, Calamito M, Joshi RP, Birnbaum MJ, Koretzky GA: AKT1 and AKT2 maintain hematopoietic stem cell function by regulating reactive oxygen species. Blood 2010, 115:4030–4038. [Full Paper][PubMed][PubMed Central]
22. Thompson BJ, Jankovic V, Gao J, Buonamici S, Vest A, Lee JM, Zavadil J, Nimer SD, Aifantis I: Control of hematopoietic stem cell quiescence by the E3 ubiquitin ligase Fbw7. J. Exp. Med. 2008, 205:1395–1408. [Full Paper][PubMed]
23. Zhang J, Grindley JC, Yin T, Jayasinghe S, He XC, Ross JT, Haug JS, Rupp D, Porter-Westpfahl KS, Wiedemann LM, et al.: PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature 2006, 441:518–522. [Full Paper][PubMed]
24. Martin-Martin N, Sutherland JD, Carracedo A: PML: Not all about tumor suppression [Internet]. Front. Oncol. 2013, 3. [Full Paper][PubMed][PubMed Central]
25. Keniry A, Oxley D, Monnier P, Kyba M, Dandolo L, Smits G, Reik W: The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat. Cell Biol. 2012, 14:659–665. [Full Paper]
26. Jeong M, Sun D, Luo M, Huang Y, Challen GA, Rodriguez B, Zhang X, Chavez L, Wang H, Hannah R, et al.: Large conserved domains of low DNA methylation maintained by Dnmt3a. Nat. Genet. 2013, doi:10.1038/ng.2836. [Full Paper][PubMed]
27. Liu B, Yee KM, Tahk S, Mackie R, Hsu C, Shuai K: PIAS1 SUMO ligase regulates the self-renewal and differentiation of hematopoietic stem cells. EMBO J. 2013, doi:10.1002/embj.201283326. [Full Paper][PubMed]
28. Weidner CI, Walenda T, Lin Q, Wölfler MM, Denecke B, Costa IG, Zenke M, Wagner W: Hematopoietic stem and progenitor cells acquire distinct DNA-hypermethylation during in vitro culture. Sci. Rep. 2013, 3:3372. [Full Paper][PubMed]
29. Cheung TH, Quach NL, Charville GW, Liu L, Park L, Edalati A, Yoo B, Hoang P, Rando TA: Maintenance of muscle stem-cell quiescence by microRNA-489. Nature 2012, 482:524–528. [Full Paper][PubMed]
30. Lechman ER, Gentner B, van Galen P, Giustacchini A, Saini M, Boccalatte FE, Hiramatsu H, Restuccia U, Bachi A, Voisin V, et al.: Attenuation of miR-126 activity expands HSC in vivo without exhaustion. Cell Stem Cell 2012, 11:799–811. [Full Paper][PubMed]
31. Crist CG, Montarras D, Buckingham M: Muscle satellite cells are primed for myogenesis but maintain quiescence with sequestration of Myf5 mRNA targeted by microRNA-31 in mRNP granules. Cell Stem Cell 2012, 11:118–126. [Full Paper][PubMed]
32. Medina R, Zaidi SK, Liu C-G, Stein JL, vanWijnen AJ, Croce CM, Stein GS: MicroRNAs 221 and 222 bypass quiescence and compromise cell survival. Cancer Res. 2008, 68:2773–2780. [Full Paper][PubMed]
33. Singh RP, Shiue K, Schomberg D, Zhou FC: Cellular epigenetic modifications of neural stem cell differentiation. Cell Transplant. 2009, 18:1197–1211. [Full Paper][PubMed][PubMed Central]
34. Evertts AG, Manning AL, Wang X, Dyson NJ, Garcia BA, Coller HA: H4K20 methylation regulates quiescence and chromatin compaction. Mol. Biol. Cell 2013, 24:3025–3037. [Full Paper][PubMed][PubMed Central]
35. Biehs B, Hu JK-H, Strauli NB, Sangiorgi E, Jung H, Heber R-P, Ho S, Goodwin AF, Dasen JS, Capecchi MR, et al.: BMI1 represses Ink4a/Arf and Hox genes to regulate stem cells in the rodent incisor. Nat. Cell Biol. 2013, 15:846–852. [Full Paper][PubMed]
36. Zacharek SJ, Fillmore CM, Lau AN, Gludish DW, Chou A, Ho JWK, Zamponi R, Gazit R, Bock C, Jäger N, et al.: Lung stem cell self-renewal relies on BMI1-dependent control of expression at imprinted loci. Cell Stem Cell 2011, 9:272–281. [Full Paper][PubMed][PubMed Central]
37. Juan AH, Derfoul A, Feng X, Ryall JG, Dell'Orso S, Pasut A, Zare H, Simone JM, Rudnicki MA, Sartorelli V: Polycomb EZH2 controls self-renewal and safeguards the transcriptional identity of skeletal muscle stem cells. Genes Dev. 2011, 25:789–794. [Full Paper][PubMed]
38. Woodhouse S, Pugazhendhi D, Brien P, Pell JM: Ezh2 maintains a key phase of muscle satellite cell expansion but does not regulate terminal differentiation. J. Cell Sci. 2013, 126:565–579. [Full Paper][PubMed]
39. So
A-Y, Jung J-W, Lee S, Kim H-S, Kang K-S: DNA methyltransferase controls stem cell aging by
regulating BMI1 and EZH2 through microRNAs. PLoS
ONE 2011, 6:e19503.
[Full Paper] ● Link between different epigenetic regulations in
the same situation
40. Lane SW, Scadden DT, Gilliland DG: The leukemic stem cell niche: current concepts and therapeutic opportunities. Blood 2009, 114:1150–1157. [Full Paper][PubMed]
41. Lang D, Mascarenhas JB, Shea CR: Melanocytes, melanocyte stem cells, and melanoma stem cells. Clin. Dermatol. 2013, 31:166–178. [Full Paper]
42. Clevers H: The cancer stem cell: premises, promises and challenges. Nat. Med. 2011, doi:10.1038/nm.2304. [Full Paper]
43. Friedman RC, Farh KK-H, Burge CB, Bartel DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19:92–105. [Full Paper][PubMed]
44. Bernstein BE, Meissner A, Lander ES: The mammalian epigenome. Cell 2007, 128:669–681. [Full Paper][PubMed]
45. Govin J, Khochbin S: Histone variants and sensing of chromatin functional states. Nucl. Austin Tex 2013, 4. [PubMed]
46. He C, Huang H, Xu L: Mechanisms guiding Polycomb activities during gene silencing in Arabidopsis thaliana. Front. Plant Sci. 2013, 4:454. [Full Paper][PubMed]
47. Lim S, Kaldis P: Cdks, cyclins and CKIs: roles beyond cell cycle regulation. Dev. Camb. Engl. 2013, 140:3079–3093. [PubMed]



