Complex splicing in the human T-cell leukemia virus (HTLV) family of retroviruses: a number of proteins play a key role in the establishment and maintenance of viral infection.
2012/10/01
Abstract
Like all retroviruses, Human T-lymphotropic virus type-1 (HTLV-1) and type 2 (HTLV-2) genomes are encoding for structural and enzymatic proteins (Gag, Pro, Pol, Env), but they also contain a pX region in their 3' end that possesses several open reading frames (ORFs). Those ORFs encode regulatory (Tax, Rex, and HBZ - HTLV-1 basic leucine zipper factor) as well as auxiliary proteins translated either from singly spliced mRNAs and coding for p12, p8, p13 (HTLV-1) and p28 (HTLV-2), or from doubly spliced mRNAs and coding for p30 (HTLV-1), p10 and p11 (HLTV-2). Here, we review the current knowledge on HTLV-1/2 auxiliary proteins' functions. Those proteins modulate viral transmission, viral persistence, host cell transcription and fate, and escape from the host immune system.
Table of Contents
- Introduction
- HTLV-1/2 Tax and Rex
- HTLV-1 p30 and HTLV-2 p28
- HTLV-1 p12 and HTLV-2 p10 and p11
- p12 activates STAT5 signaling and increases T-cell proliferation.
- p12 controls calcium release from the ER and T-cell activation.
- p12 re-routes the major histocompatibility complex (MHC) class I to the proteasome for degradation.
- p12 promotes the escape from the immune system of the host.
- p12 and the H+ vacuolar ATPase.
- HTLV-1 p8
- HTLV-1 p13
- Conclusions
- Acknowledgments
- References and recommended reading
Introduction
The Human T-lymphotropic virus (HTLV) family of complex retroviruses is composed of four members: HTLV-1, -2, -3, -4. According to phylogenetic analyses, almost all HTLVs (except HTLV-4, [1]) have a simian counterpart [2,3].
HTLV-1 is an oncovirus responsible for a peculiar type of leukemia called Adult T-cell leukemia/lymphoma (ATLL), as well as for a neurological disease named HTLV-I Associated Myelopathy/Tropical Spastic Paraparesis (HAM/TSP). On the other hand, HTLV-2, -3 -4 infection has not been linked to any cancer. We will focus our analysis on HTLV-1 and HTLV-2, which can be transmitted either in a horizontal way (sex, blood) or from the mother to the baby, notably through breast-feeding. Ten to twenty million people are infected with HTLV-1 worldwide, while 5 millions are infected with HTLV-2. Importantly, only 5% of all HTLV-1 infected persons will develop a disease, years after the initial infection. HTLV-1 and -2 target CD4+ and CD8+ T cells respectively in vivo [4]. Those retroviruses mostly expend through clonal expansion of infected cells and barely use their reverse transcriptase. HTLV-1 and HTLV-2 share approximately 70% sequence homology (Figure 1, [5]). Like all retroviruses their genomes are coding for structural and enzymatic proteins (Gag, Pro, Pol, Env). However, HTLVs specifically present in their 3' part a pX region containing several ORFs [6‑9]. Proteins encoded by those pX ORFs are obtained after complex splicing of their mRNAs followed by their translation: there are regulatory (Tax, Rex and HBZ) and auxiliary (p12/p8, p13 and p30 for HTLV-1 and p10, p11 and p28 for HTLV-2) proteins.
HTLV-1 and HTLV-2 tax/rex mRNA are highly expressed compared to those of the auxiliary proteins which are expressed 10−1000-fold lower [10]. Western blot analyses have failed to detect HTLV-1/2 auxiliary proteins in lysates from HTLV infected cell lines. This result is consistent with their low mRNA levels.
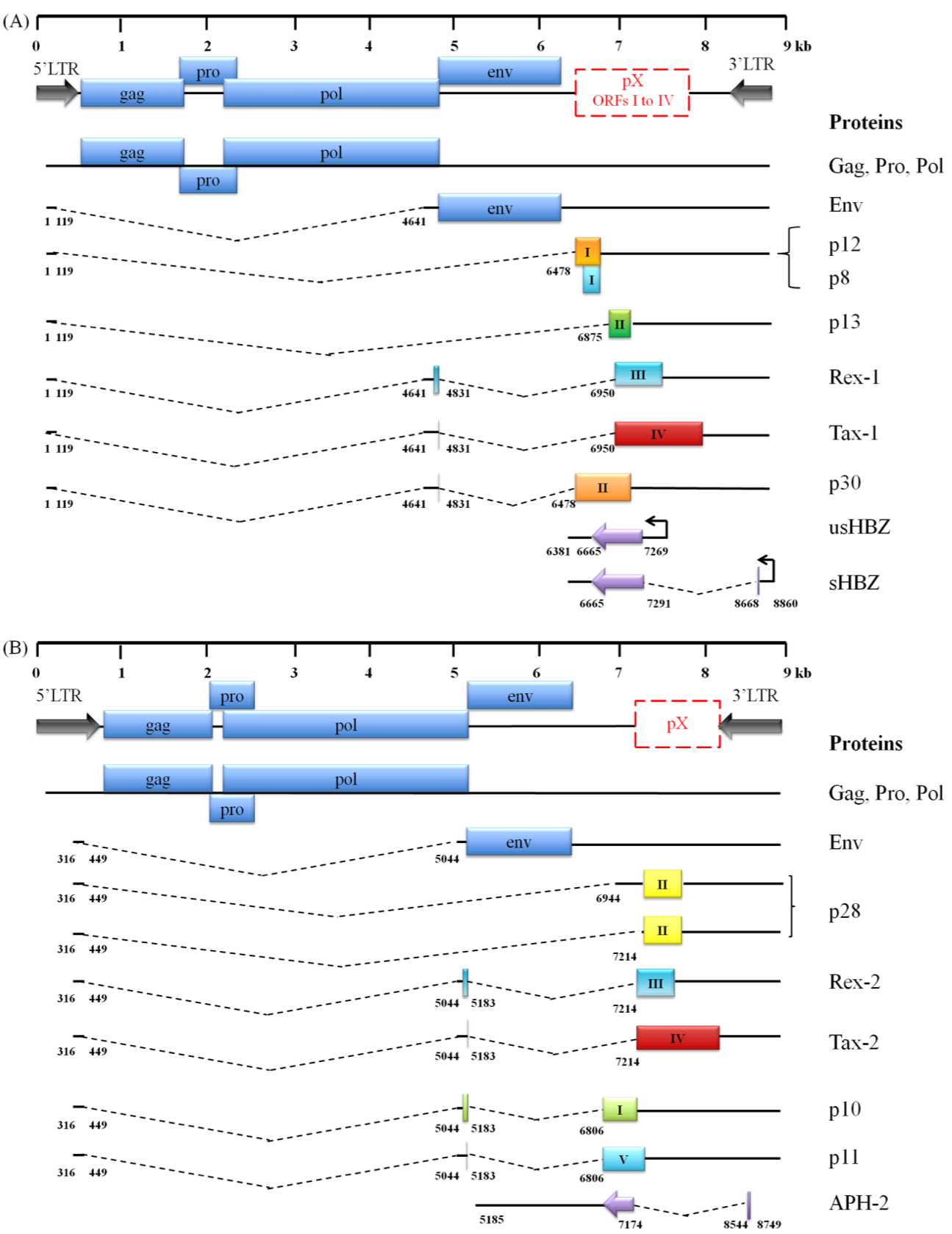
Figure 1. Comparison of the genomic organization, alternative splicing, and coding potential of HTLV-1 (A) and HTLV-2 (B) mRNAs. .
Spliced mRNAs and viral proteins are shown: HTLV-1 and HTLV-2 genomes encode common structural and enzymatic proteins found in all retroviruses (Gag, Pro, Pol and Env) and several regulatory (Tax, Rex and HBZ) and auxiliary proteins, due to a tight splicing. ORFs are indicated by colored boxes. Splice sites are indicated by numbers corresponding to nucleotides starting from the 5' end of the provirus.
HTLV-1/2 Tax and Rex
HTLV-1/2 genomes encode regulatory proteins (Tax and Rex), which regulate viral expression. Tax is a transcriptional transactivator which is required for a proper viral transcription from the 5'LTR (Long Terminal Repeat). Tax recruits the CREB (cAMP response element-binding Protein) transcription factor and co-activators such as CBP (CREB-binding protein)/p300 onto the 5'LTR (Figure 2 (2)). Tax is also essential for cell transformation. It alters the transcription of several cellular genes involved in cell growth and differentiation, cell cycle control, DNA repair or apoptotic processes [11].
Rex acts at a post-transcriptional level. It is responsible for the nuclear export of unspliced gag/pol and incompletely spliced env mRNAs into the cytoplasm. It binds to the RxRE (Rex responsive element) located on the viral mRNAs, and to the CRM-1 (chromosome maintenance region 1) cellular export factor (Figure 2 (3, 4), [12]).
HTLV-1 and HTLV-2 also produce anti-sense proteins after translation of a spliced mRNA whose promoter is located in the 3'LTR: those proteins were named HBZ (HTLV-1 basic leucine zipper factor) and APH2 (anti-sense protein of HTLV-2) respectively. hbz mRNA favors T-cell growth [13,14], while the HBZ protein inhibits Tax-mediated transcription [15], thus decreasing viral expression.
Herein, we will review the current knowledge of HTLV-1 and HTLV-2 auxiliary proteins in vitro.
HTLV-1 p30 and HTLV-2 p28
p30 and p28: negative regulators of both Tax and Rex expression.
HTLV-1 p30 is translated from a doubly spliced mRNA transcribed from ORF II. p30 is located within nucleoli and nuclei [8,16] (Figure 2 (5)). Instead of Rex, p30 is not able to shuttle between the nucleus and the cytoplasm: indeed p30 harbors two nucleolar retention signals (NoRS), and three nuclear localization sequences (NLS) [12,17,18]. It also contains a Rex-binding domain, a p300-binding domain and a DNA-binding domain [19].
p30 specifically sequesters tax/rex mRNA in the nucleus
p30 is a negative regulator of viral replication. At the post-transcriptional level, it counteracts the positive effects of Tax and Rex. In the nucleus, it specifically binds to and retains the doubly spliced tax/rex mRNA, resulting in a decreased expression of those regulatory proteins [12]. Other results suggest that p30 binds to the tax/rex mRNA splice junction [20,21], explaining the specific retention of those mRNAs (Figure 2 (6)). Thus, by down-regulating viral production, p30 promotes viral latency.
The negative feedback loop between Rex and p30
Within nucleoli of infected cells, p30 also directly interacts with Rex through its RNA binding domain, preventing Rex from binding to RxRE (Rex responsive element) (Figure 2 (7)). Nevertheless, this association depends upon the level of expression of the two proteins: during early infection Rex is highly produced and thus exerts its export activity by binding onto RxRE. In contrast, when p30 is in excess within the nucleus, it binds to Rex, preventing its association with RxRE, thus inhibiting viral mRNAs export to the cytoplasm [22,23]. For more details on this interaction please refer to [22]. It has also been demonstrated [24] that p30 is able to sequester Rex/CRM-1 complexes in nucleoli, thus preventing their association in the nucleoplasm with mRNA that should be exported (Figure 2 (8)).
p30 and Tax-mediated transcription
p30 also interacts with CBP/p300 proteins [25,26] that are essential for activating viral transcription (see above and Figure 2 (9)). This prevents the association between CBP/p300 and Tax, and in turn decreases Tax-mediated viral transcription. Thus, by interacting with CBP/p300, p30 acts as a transcriptional regulator in the infected cells.
Similarly, it was stressed that the HTLV-2 ORF II protein, i.e. p28, in addition to its localization to the nucleus and probably because it shares identity with HTLV-1 p30, also sequesters tax/rex mRNA in the nucleus [21]. Therefore, both p30 and p28 down-modulate viral expression, alter the expression of the two regulatory proteins (Tax and Rex) and, in turn, promote viral persistence by preventing those proteins from being detectable by the host immune system. However, p30 and p28 HTLV proteins use two distinct mechanisms: while HTLV-1 p30 specifically binds to and retains tax/rex mRNA, HTLV-2 p28 inhibits its export by counteracting the TAP (Tip-associated protein)/p15 export pathway (Figure 2 (10), [27]). Besides, unlike p30, p28 has no transcriptional repressive activity, as it does not interfere with Tax-mediated transcription, but only acts as a repressor at the post-transcriptional level.
p30 interferes with TLR4 signaling.
Toll-like Receptor 4 (TLR4) is a Pathogen Recognition Receptor (PRR), located at the cell surface. When TLR4 recognizes its ligand, it triggers a signaling cascade leading to the production of pro-inflammatory cytokines and type I interferon to insure a proper and fast innate immune response. p30 contributes to the down-regulation of TLR4 expression at the cell surface and decreases pro-inflammatory cytokines production by infected T-cells. Moreover, upon TLR4 stimulation by LPS (lipopolysaccharides) in human macrophages [28], p30 increases GSK3β kinase phosphorylation, resulting in its inactivation, which is a key step for interleukin-10 (IL-10) production. Therefore, p30 promotes a reduction of the immune response against infected viral protein cells and thus favors viral persistence in the host.
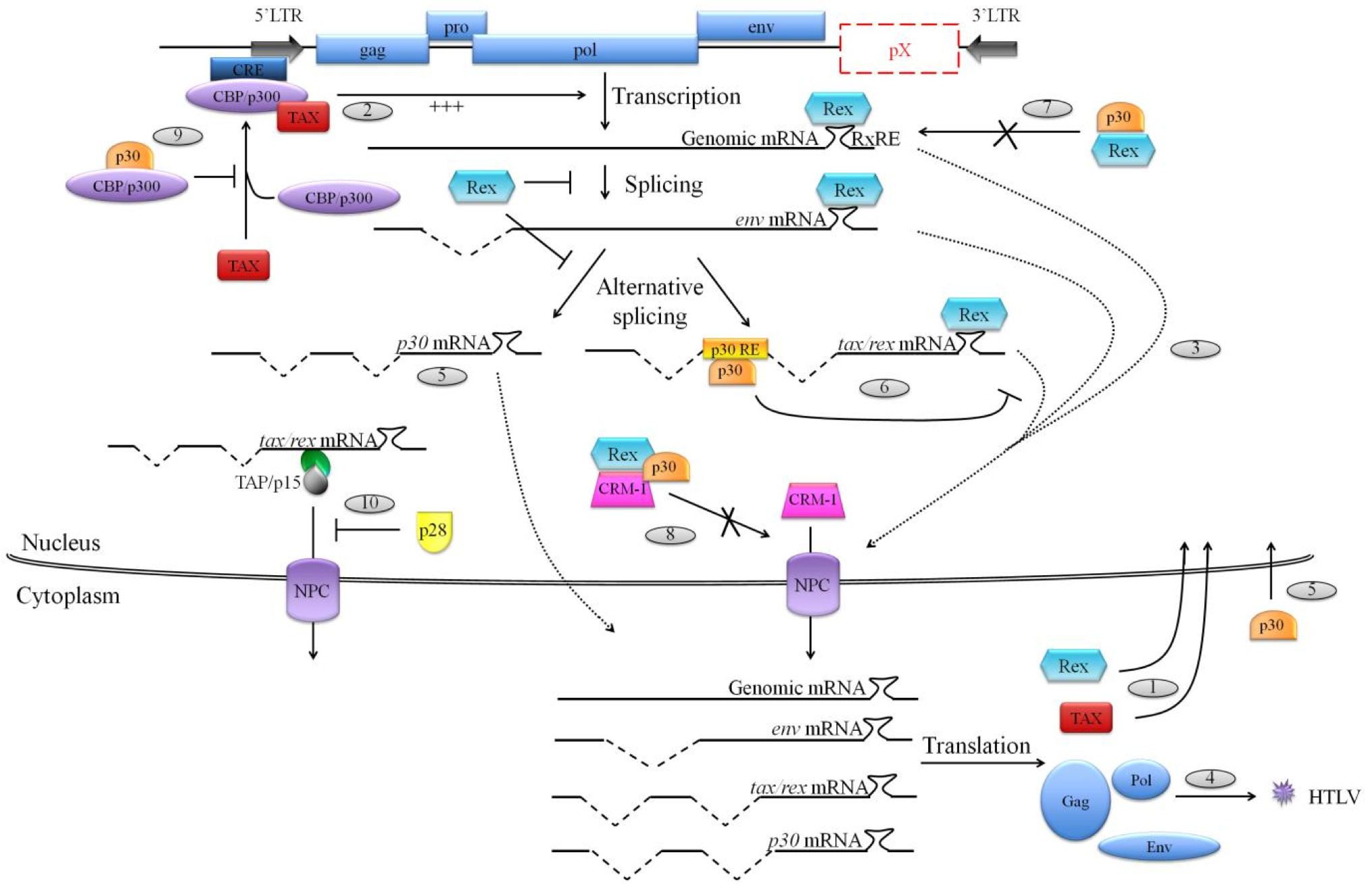
Figure 2. Functions of HTLV-1 p30 and HTLV-2 p28 on virus replication.
(1) Alternatively double spliced tax/rex mRNA is the first to be exported to the cytoplasm in order to be translated into the Tax and Rex regulatory proteins, which will translocate to the nucleus. (2) Tax is a transcriptional transactivator of HTLV promoter and is required for a proper transcription of viral genes from the 5'LTR. (3) Meanwhile, on the one hand Rex inhibits splicing processes and on the other hand it binds to Rex responsive element (RxRE) located on viral mRNAs, and also binds to the cellular export factor CRM-1 in order to export those viral mRNAs to the cytoplasm for their translation and (4) the production of new virions. (5) HTLV-1 p30 is translated from a doubly spliced mRNA transcribed from ORF II, and HTLV-2 p28 is translated from a singly spliced mRNA transcribed from ORF II. Once synthesized, those two proteins localize to the nucleus. (6) In the nucleus, p30 specifically sequesters tax/rex mRNA by binding to p30 responsive element (p30 RE) located at the tax/rex mRNA splice junction. (7) Moreover, p30 can directly bind to Rex, preventing its binding to the RxRE and also (8) interacts with Rex/CRM-1 complexes to inhibit their association with viral mRNAs to export. (9) Furthermore, p30 negatively interferes with CBP/p300 association with Tax, preventing an association that is essential for a strong activation of the viral 5'LTR. (10) Finally, HTLV-2 p28 also retains tax/rex mRNA in the nucleus. Nevertheless, this sequestration is mediated by the inhibition of the TAP/p15 export pathway.
HTLV-1 p12 and HTLV-2 p10 and p11
HTLV-1 p12 is translated from a singly spliced mRNA transcribed from ORF I [8,9]. Ectopically expressed p12 was found in the cellular endomembranes, and particularly in the perinuclear area [16]. p12 accumulates in the endoplasmic reticulum (ER) and the cis-Golgi apparatus. The protein is composed of two putative transmembrane domains, four putative proline-rich (PXXP) Src homology 3 (SH3)-binding domains, two putative leucine zipper (LZ) motifs, a putative adaptin motif, and a non-canonical ER retention/retrieval motif at its N- terminus [6].
p12 activates STAT5 signaling and increases T-cell proliferation.
In ER, p12 binds to the immature form of the interleukin-2 receptor (IL-2R) γ and β chains (Figure 3 (1)). Sequestration of IL-2R and its accumulation in the ER entails the phosphorylation of STAT5 (Signal Transducer and Activator of Transcription 5). Under normal conditions, after interleukin-2 (IL-2) binds to its receptor, IL-2R is activated and its γ and β chains activate the JAK (Janus kinase)/STAT pathway. This results in STATs phosphorylation entailing their dimerization and the entry in the nucleus of this dimer to act as a transcription factor.STAT5 activation induces IL-2 expression. IL-2 stimulates T-cells proliferation and differentiation. Thus, aggregation of IL-2R in the ER, mimics the phosphorylation of STAT5 induced by IL-2 signaling and, in turn, increases T-cell proliferation by lowering the threshold of IL-2 requirement for T-cell growth [29].
p12 controls calcium release from the ER and T-cell activation.
ER is the reservoir of calcium ions (Ca2+) in the cell. Thus, p12 localization might suggest an effect on the regulation of intracellular calcium concentration. p12 binds to calreticulin and calnexin, two chaperone proteins, that ensure appropriate protein folding and play a role in Ca2+ storage in the ER. This entails Ca2+ release from the ER and activation of the transcription factor NFAT (Nuclear factor of activated T- cells). Indeed, in the cytoplasm calcium ions interact with calmodulin/calcineurin complexes promoting dephosphorylation of the cytoplasmic inactive NFAT. Once dephosphorylated, the transcription factor migrates within the nucleus to induce IL-2 gene expression (Figure 3 (2), [30]). Thereby, by increasing cytoplasmic Ca2+ concentration in infected T-cells, p12 promotes IL-2 production and in turn, T-cell proliferation [6].
p12 re-routes the major histocompatibility complex (MHC) class I to the proteasome for degradation.
MHC class I molecules (MHC-I) biosynthesis occurs in ER. MHC-I α heavy chain is associated with calnexin before it associates with β2-microglobulin to form mature MHC-I molecules which will be secreted at the cell surface to present antigen to cytotoxic T lymphocytes (CTLs) and thus activate them. p12 interacts with the heavy MHC-I chain preventing its association with the β2-microglobulin, and leads to its degradation by the proteasome (Figure 3 (3), [31]). Thus, this leads to a decrease in MHC-I expression at the cell surface and in turn promotes the escape of HTLV-1 infected cells from the immune surveillance.
p12 promotes the escape from the immune system of the host.
Decrease in MHC-I expression from the surface of infected T-cells could activate the innate immune response, as natural killer (NK) cells, in the absence of an inhibitory signal provided by MHC-I recognition, receive an activating signal which leads to lysis of the abnormal cell. p12 lowers the expression of NK cells activating ligand ICAM (Intercellular Adhesion Molecule) at the surface of the infected cell (Figure 3 (4), [32]). Consequently, NK cells do not detect HTLV-1 infected cells, promoting a bypass of the immune surveillance and the persistence of the viral infection.
p12 and the H+ vacuolar ATPase.
The H+ vacuolar ATPase (V-ATPase) is a proton pump needed for the acidification of various intracellular compartments such as ER, lysosomes, endosomes, Golgi vesicles or clathrin coated vesicles. p12 interacts with the 16 kDa protein of the V-ATPase, and may interfere with its function. Several processes depend on acidification: secretory pathway, endocytic pathway, biosynthesis of MHC-II-antigen complexes in endosomes [33]. Thereby, p12 interfering with the synthesis and secretion of MHC-II-antigen complexes is another example of how to bypass activation of the immune system by an HTLV-1 protein.
HTLV-2 p10 and p11 present functional analogy with p12 since both bind to MHC-I heavy α chain. However, they do not bind other p12 targets, such as the IL-2R β chain or the 16-kDa subunit of the vacuolar H+ ATPase. Moreover, HTLV-1 p12 and HTLV-2 p10 and p11 present different cellular localizations. Indeed, p10 and p11 localize in the nucleus of infected cells [34].
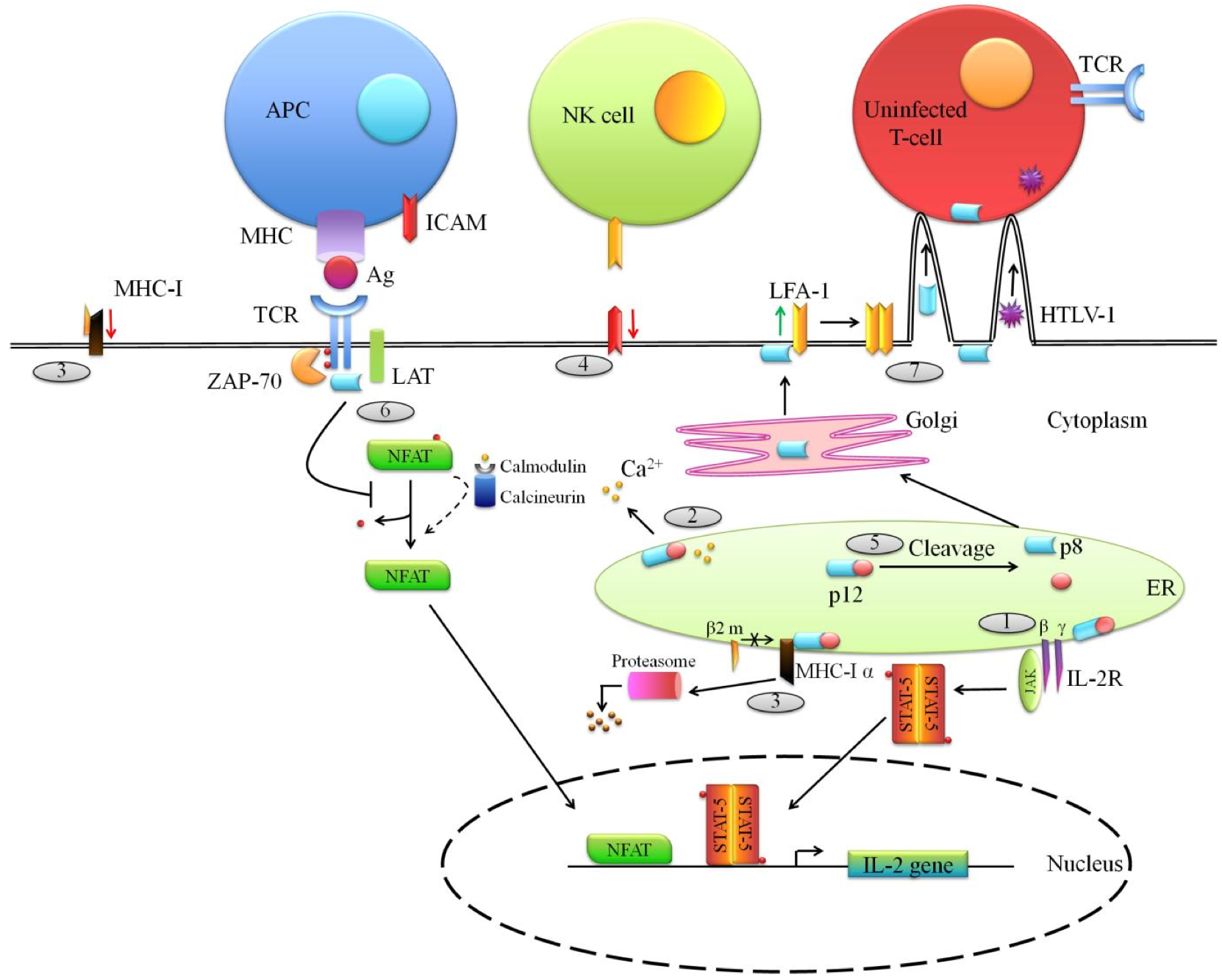
Figure 3. Functions of the ORF-I proteins (p12 and p8) of HTLV-1. .
HTLV-1 p12 is translated from a singly spliced mRNA transcribed from ORF I. (1) In the endoplasmic reticulum (ER), p12 binds to and retains the immature form of the IL-2 receptor (IL-2R) γ and β chains and this accumulation in the ER entails the phosphorylation of the Signal Transducer and Activator of Transcription 5 (STAT5) which is translocated to the nucleus to promote IL-2 expression. (2) In addition, p12 mediates calcium ions release from the ER. Ca2+ interacts with calmodulin/calcineurin complexes promoting dephosphorylation of the cytoplasmic inactive NFAT (Nuclear factor of activated T-cells) which migrates within the nucleus to induce IL-2 gene expression. (3) Furthermore, within the ER, p12 interacts with the heavy α chain of MHC-I molecules preventing its association with the β2-microglobulin (β2 m) and leads to its degradation by the proteasome. Thus, it entails a decrease of MHC-I expression at the cell surface. MHC-I down-regulation expression could promote NK cells activation. To avoid this, (4) p12 lowers the expression of NK cells activating ligand ICAM (Intercellular Adhesion Molecule) at the surface of the infected cell. (5) The p12 protein can be cleaved in the ER, leading to the formation of p8 which then traffics to the cell surface. (6) There, p8 negatively interferes with TCR signaling by decreasing LAT phosphorylation resulting in a down-regulation of NFAT activity. Here are depicted two opposite effects of HTLV-1 auxiliary proteins (see above (2)). (7) At the cell surface, p8 also modulates the adhesion molecule LFA-1 clustering at the cell surface, promoting the formation of cell-to-cell contacts through which virions and even p8 are transmitted to uninfected cells.
HTLV-1 p8
p8 inhibits the T-cell receptor (TCR) signalization, leading to T-cell anergy.
When an antigen carried by an antigen- presenting cell (APC) is recognized by the TCR of a given T-cell, TCR signalization is engaged. This consists in a cascade of phosphorylation events and leads to activation of NF-κB or NFAT transcription factors, in order to enhance T-cell proliferation and differentiation. Proteolytic cleavage of p12 amino terminus in the endoplasmic reticulum (ER) results in the formation of p8 which then traffics to the cell surface (Figure 3 (5)). Interestingly, p8 is recruited to the site of interaction between an APC and a T-cell, called the immunological synapse. There, p8 down-regulates TCR signaling and leads to T-cell anergy in a LAT (linker of activated T cells)-dependent manner. p8 decreases LAT phosphorylation resulting in a down-regulation of NFAT activity, and thus inhibits the proliferation and differentiation of T-cells (Figure 3 (6)). Therefore, p12 and p8 have opposite effects: p12 tends to accelerate T-cell proliferation and differentiation in a calcium-dependent manner, while p8 down-regulates those events when T-cells are activated [19,33].
p8 increases the formation of cellular conduits, promoting viral transmission.
HTLV-1 transmission is more efficient through cell-to-cell contact. Interestingly, p8 modulates LFA-1 (lymphocyte function-associated antigen 1) adhesion molecule clustering at the cell surface, thus promoting formation of cell-to-cell contacts and thus favoring virus transmission (Figure 3 (7)). p8 increases number and length of inter-cellular conduits [35], promoting communication between infected and uninfected cells target, thus facilitating viral transmission. p8 is also transferred to uninfected cells through those conduits, allowing to form new conduits and to accelerate HTLV-1 transmission. The transmission of viruses using those cellular conduits might also allow them to avoid their possible recognition by the immune system. Even if HTLV-2 also persists in its hosts and escape from the immune surveillance, no HTLV- 2 ortholog of HTLV-1 p8 has been identified so far.
HTLV-1 p13
Encoded by ORFII the p13 protein corresponds to the C-terminal 87 amino acids of p30 [36]. Thus, p13 does not have the nuclear localization sequence (NLS) of p30, and accumulates in the inner membrane of mitochondria. Initially p13 was however considered as a nuclear protein: indeed, Koralnik et al. [16] have stressed that upon p13 transfection in HeLa cells, the protein localized within the nucleus. This might result from p13 overexpression. HTLV-1 p13 is composed of several domains [36]: an amphipathic alpha helix containing a mitochondrial targeting sequence (MTS), a transmembrane region, a region with a high flexibility score and a C-terminal region. This latter region (the C-terminal) includes PXXP motifs that may allow the interaction between p13 and proteins containing SH3 domains. It also contains a NLS that is probably responsible of the nuclear targeting of p13 when the MTS is masked. The mitochondrial localization of HTLV-1 p13 suggests that it impacts mitochondrial functions, while its nuclear localization supposes effects on transcriptional regulation.
Until now, no functional homologue of p13 has been described in the HTLV-2 genome.
p13 and the regulation of Tax-mediated transcription in the nucleus.
A recent report [37] stressed that, when highly expressed or when Tax is co-expressed, p13 is localized within the nucleus. In fact, Tax mediates p13 ubiquitination, stabilizing the protein which is then partially re-localized to the nucleus. There, p13 bind to Tax, preventing the interaction between Tax and CBP/p300 (Figure 4 (1)). As a result, p13, like p30, inhibits Tax-mediated viral transcription, decreasing HTLV-1 expression, and hence contributes to viral latency and viral persistence in vivo.
p13 impairs mitochondrial functions:
p13 increases mitochondrial K+ permeability and thus favors apoptosis
Cytochrome c is a component of the electron transport chain in mitochondria, and acts as a pro-apoptotic signal: when released in the cytoplasm it triggers the caspase cascade leading to apoptosis. In mitochondria, p13 alters the inner mitochondrial membrane potential to stimulate K+ influx by the organelle and cause Cytochrome c is a component of the electron transport chain in mitochondria, and acts as a pro-apoptotic signal: when released in the cytoplasm it triggers the caspase cascade leading to apoptosis. In mitochondria, p13 alters the inner mitochondrial membrane potential to stimulate K+ influx by the organelle and cause
p13 increases mitochondrial ROS production
p13-induced K+ influx, activation of the electron transport chain and mitochondrial depolarization promotes mitochondrial respiration, increasing O2 consumption. This favors ROS (reactive oxygen species) production by mitochondria (Figure 4 (4)), which have distinct effects according to cell state. Actually, ROS entail activation of resting cells, while in transformed cells they trigger the programmed cell death. p13 function may contribute to HTLV-1 persistence since, by modulating ROS production, p13 lowers the number of transformed cells that could be recognized by the immune system of the host. Thus, controlling ROS production is another way for p13 to bypass the immune recognition and to promote viral persistence. For more detailed information on this process, please refer to this study [38].
p13 decreases mitochondrial Ca2+ uptake
Changes in the inner mitochondrial membrane potential are involved in the reduction of mitochondrial Ca2+ uptake (Figure 4 (5)). However, it is unlikely that this consequence of p13 expression modulates Ca2+ signaling in infected cells or favors expression of genes controlled by Ca2+-regulated transcription factors, such as NFAT, since the reduction of Ca2+ uptake mediated by p13 seems to alter cytosolic Ca2+ concentration only locally [39]. Ultimately and like p30, p13 protein promotes viral latency, by decreasing Tax-mediated transcription; and alters cell fate (apoptosis of transformed cells), thus contributing to the bypass of the immune surveillance and in turn favors viral persistence.
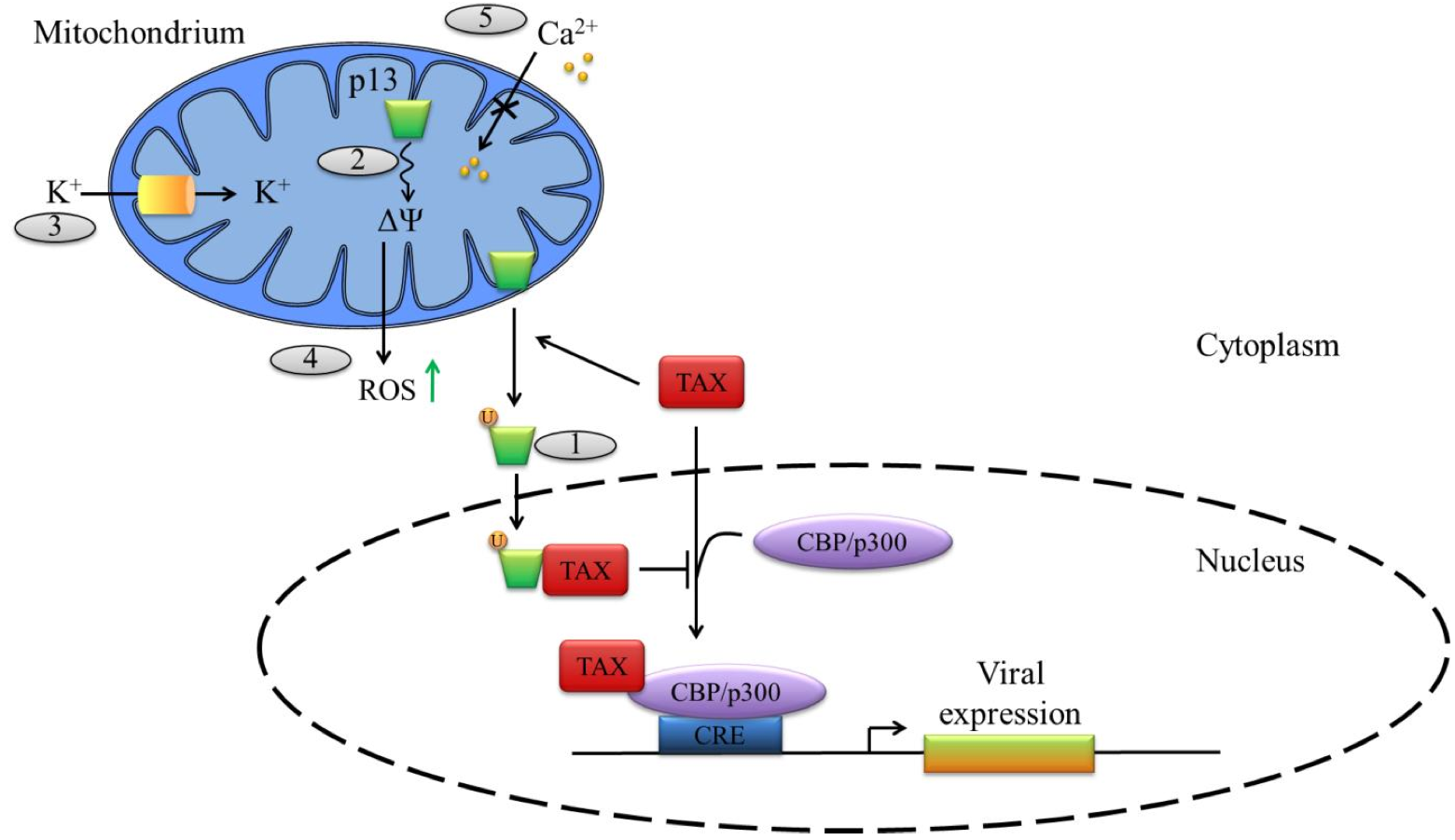
Figure 4. Functions of the ORF-II p13 of HTLV-1. .
HTLV-1 p13 is translated from a singly spliced mRNA transcribed from ORF II and is localized at the mitochondrial inner membranes. (1) In the cytoplasm, Tax mediates p13 ubiquitination, stabilizing the protein, which is then partially re-localized to the nucleus where it can bind to Tax and prevents its association with the transcriptional co-activating proteins CBP/p300. (2) p13 alters the inner mitochondrial membrane potential (ΔΨ) and thus (3) increases mitochondrial K+ permeability as well as (4) ROS production – which can favors apoptosis – and (5) decreases calcium uptake by the mitochodrium.
Conclusions
Despite the limited size of their genome, complex retroviruses like HTLVs but also BLV (Bovine leukemia virus) and HIV (Human immunodeficiency virus) have evolved several strategies to synthesize their structural and enzymatic proteins (Gag, Pol, Env), as well as regulatory and auxiliary proteins (Figure 1).
Throughout this review, we have highlighted the role of HTLV-1 and HLTV-2 auxiliary proteins (Figure 2, 3 and 4; Table 1) in the establishment of a lifelong viral persistence in host cells. Those auxiliary proteins confer lots of advantages to those viruses. Indeed, they favor viral transmission, modulate transcription and promote viral latency. In addition, those proteins also contribute to evasion from the immune system of the host by reducing the activation of CTLs and NK cells or by inducing programmed cell death. Finally they alter cellular signalization and in turn impact cellular fate and mediate cell proliferation and differentiation.
Evenif HTLV-1 and HTLV-2 share approximately 70% genomic homology from a structural point of view, they present different pathogenic potential. There are still remaining questions concerning the differences in pathogenicity of the two viruses, and one can wonder if those differences can be explain by the function of their auxiliary proteins.
Therefore, it is very likely that understanding the function of HTLV-1/2 auxiliary proteins, such as the rate of viral mRNAs expression [10], the cellular localization of the proteins and their functions, might give us insight into the mechanisms that drive virus latency and evasion from host immune detection.
Some HTLV-1 auxiliary proteins are expressed in vivo [40], thus, further investigations concerning the function of those proteins might be useful to uncover targets for new potential therapeutics. Indeed, as regards HIV-1, the study of its auxiliary proteins (Vpu, Vpr, Vif, and Nef proteins) enabled researchers to decipher the infection process [41]. Similarly in-depth studies of HTLV auxiliary proteins may improve our understanding of HTLV transmission, viral latency or escape from the host immune system.
Table 1. Functional characteristics of HTLV-1 and HTLV-2 auxiliary proteins.
Protein | Localization | Function(s) | References |
HTLV-1 p30 | Nucleolus, nucleus | sequestrates tax/rex mRNA in the nucleus; binds to Rex thus inhibiting viral mRNAs export to the cytoplasm; decreases Tax-mediated viral transcription; interferes with TLR4 signaling. | 12; 22; 26; 28 |
HTLV-2 p28 | nucleus | sequestrates tax/rex mRNA in the nucleus | 21 |
HTLV-1 p12 | Endoplasmic reticulum, cis-Golgi apparatus | binds to the IL-2R γ and β chains and thus activates STAT5 signaling; interacts with calreticulin and calnexin promoting Ca2+ release from the ER and NFAT activation; leads to the degradation of the heavy α chain of MCH‑I molecules by the proteasome; decreases ICAM expression at the cell surface. | 29 - 32 |
HTLV-2 p10 and p11 | nucleus | associate with MHC-I heavy α chain. | 6 |
HTLV-1 p8 | cell surface, immunological synapse | recruited to the immunological synapse; down-regulates TCR signaling; promotes cell-to-cell contacts through synapse LFA-1; increases the formation of cellular conduits. | 33; 35 |
HTLV-1 p13 | Mitochondrial inner membrane, nucleus | decreases Tax-mediated viral transcription in the nucleus; alters the inner mitochondrial membrane and increases mitochondrial K+ permeability; favors mitochondiral ROS production thus activating resting cells, while triggering the programmed cell death in transformed cells; decreases mitochondrial Ca2+ uptake | 36 - 39 |
Acknowledgments
I thank Dr Renaud Mahieux and Jocelyn Turpin for their help and their advices concerning this review.
References and recommended reading
Papers of particular interest have been highlighted as:
● of special interest
●● of outstanding interest
1.Wolfe ND, Heneine W, Carr JK, Garcia AD, Shanmugam V, Tamoufe U, Torimiro JN, Prosser AT, Lebreton M, Mpoudi-Ngole E, et al.: Emergence of unique primate T- lymphotropic viruses among central African bushmeat hunters.Proc. Natl. Acad. Sci. U.S.A. 2005, 102:7994–7999.
2.Koralnik IJ, Boeri E, Saxinger WC, Monico AL, Fullen J, Gessain A, Guo HG, Gallo RC, Markham P, Kalyanaraman V: Phylogenetic associations of human and simian T-cell leukemia/lymphotropic virus type I strains: evidence for interspecies transmission.J. Virol. 1994, 68:2693–2707.
3.Slattery JP, Franchini G, Gessain A: Genomic evolution, patterns of global dissemination, and interspecies transmission of human and simian T-cell leukemia/lymphotropic viruses.Genome Res. 1999, 9:525–540.
4.Kannian P, Yin H, Doueiri R, Lairmore MD, Fernandez S, Green PL: Distinct transformation tropism exhibited by human T lymphotropic virus type 1 (HTLV-1) and HTLV-2 is the result of postinfection T cell clonal expansion.J. Virol. 2012, 86:3757– 3766.
5.Rende F, Cavallari I, Romanelli MG,
Diani E, Bertazzoni U, Ciminale V: Comparison of the Genetic
Organization, Expression Strategies and Oncogenic Potential of HTLV-1 and
HTLV-2.Leukemia Research and Treatment 2012, 2012:1–14. ● This review collects information concerning the differences between
HTLV-1 and HTLV-2 which can be responsible for their distinct pathogenicity:
their genomic organization, their expression profile, and the role of their
proteins.
6.Albrecht B, Lairmore MD: Critical role of human T-lymphotropic virus type 1 accessory proteins in viral replication and pathogenesis.Microbiol. Mol. Biol. Rev. 2002, 66:396–406, table of contents.
7.Berneman ZN, Gartenhaus RB, Reitz MS Jr, Blattner WA, Manns A, Hanchard B, Ikehara O, Gallo RC, Klotman ME: Expression of alternatively spliced human T-lymphotropic virus type I pX mRNA in infected cell lines and in primary uncultured cells from patients with adult T-cell leukemia/lymphoma and healthy carriers.Proc. Natl. Acad. Sci. U.S.A. 1992, 89:3005– 3009.
8.Ciminale V, Pavlakis GN, Derse D, Cunningham CP, Felber BK: Complex splicing in the human T-cell leukemia virus (HTLV) family of retroviruses: novel mRNAs and proteins produced by HTLV type I.J. Virol. 1992, 66:1737–1745.
9.Koralnik IJ, Gessain A, Klotman ME, Lo Monico A, Berneman ZN, Franchini G: Protein isoforms encoded by the pX region of human T-cell leukemia/lymphotropic virus type I.Proc. Natl. Acad. Sci. U.S.A. 1992, 89:8813– 8817.
10.Li M, Green PL: Detection and quantitation of HTLV-1 and HTLV-2 mRNA species by
real-time RT-PCR.J. Virol. Methods 2007, 142:159–168. ●● In this study, the authors present a method to quantify specifically all
the viral mRNAs from both HTLV-1 and HTLV-2. This methodology can be used to
follow viral gene expression during the infection process, which can be useful
to decipher the possible contribution of viral proteins to viral persistence and
pathogenesis, among others.
11.Journo C, Douceron E, Mahieux R: HTLV gene regulation: because size matters, transcription is not enough.Future Microbiol 2009, 4:425–440.
12.Baydoun HH, Bellon M, Nicot C:
HTLV-1 Yin and Yang: Rex and p30 master regulators of viral
mRNA trafficking.AIDS Rev 2008, 10:195–204. ●● This review emphasises the complex mutual regulation between Rex and
p30, concerning viral mRNA nuclear export, which governs the switch between
virus latency and replication.
13.Arnold J, Zimmerman B, Li M, Lairmore MD, Green PL: Human T-cell leukemia virus type- 1 antisense-encoded gene, Hbz, promotes T- lymphocyte proliferation.Blood 2008, 112:3788–3797.
14.Satou Y, Yasunaga J, Yoshida M, Matsuoka M: HTLV-I basic leucine zipper factor gene mRNA supports proliferation of adult T cell leukemia cells.Proc. Natl. Acad. Sci. U.S.A. 2006, 103:720–725.
15.Clerc I, Polakowski N, André-Arpin C, Cook P, Barbeau B, Mesnard J-M, Lemasson I: An interaction between the human T cell leukemia virus type 1 basic leucine zipper factor (HBZ) and the KIX domain of p300/CBP contributes to the down- regulationoftax-dependentviral transcription by HBZ.J. Biol. Chem. 2008, 283:23903–23913.
16.Koralnik IJ, Fullen J, Franchini G: The p12I, p13II, and p30II proteins encoded by human T-cell leukemia/lymphotropic virus type I open reading frames I and II are localized in three different cellular compartments.J. Virol. 1993, 67:2360–2366.
17.Ghorbel S, Sinha-Datta U, Dundr M, Brown M, Franchini G, Nicot C: Human T-cell leukemia virus type I p30 nuclear/nucleolar retention is mediated through interactions with RNA and a constituent of the 60 S ribosomal subunit.J. Biol. Chem. 2006, 281:37150– 37158.
18.Nicot C, Harrod RL, Ciminale V, Franchini G: Human T-cell leukemia/lymphoma virus type 1 nonstructural genes and their functions.Oncogene 2005, 24:6026–6034.
19.Edwards D, Fenizia C, Gold H, de Castro- Amarante MF, Buchmann C, Pise-Masison CA, Franchini G: Orf-I and orf-II-encoded proteins inHTLV-1 infection and persistence.Viruses 2011, 3:861–885.
20.Nicot C, Dundr M, Johnson JM, Fullen JR, Alonzo N, Fukumoto R, Princler GL, Derse D, Misteli T, Franchini G: HTLV-1-encoded p30II is a post-transcriptional negative regulator of viral replication.Nat. Med. 2004, 10:197–201.
21.Younis I, Khair L, Dundr M, Lairmore MD, Franchini G, Green PL: Repression of human T-cell leukemia virus type 1 and type 2 replication by a viral mRNA-encoded post-transcriptional regulator.J. Virol. 2004, 78:11077–11083.
22.Sinha-Datta U, Datta A, Ghorbel S, Dodon MD, Nicot C: Human T-cell lymphotrophic virus type I rex and p30 interactions govern the switch between virus latency and replication.J. Biol. Chem. 2007, 282:14608–14615.
23.Bai XT, Baydoun HH, Nicot C:
HTLV-I p30: a versatile protein modulating virus replication
and pathogenesis.Mol. Aspects Med. 2010, 31:344–349. ● In this review, the authors gather the current knowledge concerning
HTLV-1 p30 functions in the infected cells; ranging from its role in the virus
life cycle, to its possible role in the establishment of leukemia.
24.Baydoun H, Duc-Dodon M, Lebrun S, Gazzolo L, Bex F: Regulation of the human T-cell leukemia virus gene expression depends on the localization of regulatory proteins Tax, Rex and p30II in specific nuclear subdomains. Gene 2007, 386:191–201.
25.Zhang W, Nisbet JW, Bartoe JT, Ding W, Lairmore MD: Human T-lymphotropic virus type 1 p30(II) functions as a transcription factor and differentially modulates CREB- responsive promoters. J. Virol. 2000, 74:11270–11277.
26.Zhang W, Nisbet JW, Albrecht B, Ding W, Kashanchi F, Bartoe JT, Lairmore MD: Human T-lymphotropic virus type 1 p30(II) regulates gene transcription by binding CREB binding protein/p300.J. Virol. 2001, 75:9885–9895.
27.Younis I, Boris-Lawrie K, Green PL: Human T-cell leukemia virus open reading frame II encodes a posttranscriptional repressor that is recruited at the level of transcription.J. Virol. 2006, 80:181–191.
28.Datta A, Sinha-Datta U, Dhillon NK, Buch S, Nicot C: The HTLV-I p30 interferes with TLR4 signaling and modulates the release of pro- and anti-inflammatory cytokines from human macrophages.J. Biol. Chem. 2006, 281:23414–23424.
29.Nicot C, Mulloy JC, Ferrari MG, Johnson JM, Fu K, Fukumoto R, Trovato R, Fullen J, Leonard WJ, Franchini G: HTLV-1 p12(I) protein enhances STAT5 activation and decreases the interleukin-2 requirement for proliferation of primary human peripheral blood mononuclear cells.Blood 2001, 98:823– 829.
30.Ding W, Albrecht B, Kelley RE, Muthusamy N, Kim S-J, Altschuld RA, Lairmore MD: Human T-cell lymphotropic virus type 1 p12(I) expression increases cytoplasmic calcium to enhance the activation of nuclear factor of activated T cells.J. Virol. 2002, 76:10374– 10382.
31.Johnson JM, Nicot C, Fullen J, Ciminale V, Casareto L, Mulloy JC, Jacobson S, Franchini G: Free major histocompatibility complex class I heavy chain is preferentially targeted for degradation by human T-cell leukemia/lymphotropic virus type 1 p12(I) protein.J. Virol. 2001, 75:6086–6094.
32.Banerjee P, Feuer G, Barker E: Human T-cell leukemia virus type 1 (HTLV-1) p12I down- modulates ICAM-1 and -2 and reduces adherence of natural killer cells, thereby protecting HTLV-1-infected primary CD4+ T cells from autologous natural killer cell- mediated cytotoxicity despite the reduction of major histocompatibility complex class I molecules on infected cells.J. Virol. 2007, 81:9707–9717.
33.Van Prooyen N, Andresen V, Gold H,
Bialuk I, Pise-Masison C, Franchini G: Hijacking the T- cell
communication network by the human T-cell leukemia/lymphoma virus type 1 (HTLV-1)
p12 and p8 proteins.Mol. Aspects Med. 2010, 31:333–343. ● This review describes the main functions of the HTLV-1 Orf -I products,
namely p12 and p8, which are similarly very important for HTLV-1 persistence in
the host.
34.Ciminale V, D'Agostino DM, Zotti L, Franchini G, Felber BK, Chieco-Bianchi L: Expression and characterization of proteins produced by mRNAs spliced into the X region of the human T-cell leukemia/lymphotropic virus type II.Virology 1995, 209:445–456.
35.Van Prooyen N, Gold H, Andresen
V, Schwartz O, Jones K, Ruscetti F, Lockett S, Gudla P, Venzon D, Franchini G: Human T-cell leukemia virus type 1 p8 protein increases cellular
conduits and virus transmission.Proc. Natl. Acad. Sci. U.S.A. 2010, 107:20738– 20743. ●● This paper highlights a novel mechanism, driven by HTLV- 1 p8, which
promotes viral transmission between cells. Indeed, p8 increases the number and
the length of intracellular conduits between T-cells, through which HTLV-1
virions, as well as p8 itself, are transferred to other cells, avoiding a
possible recognition by the immune system of the host.
36.Silic-Benussi M, Biasiotto R, Andresen V, Franchini G, D'Agostino DM, Ciminale V: HTLV-1 p13, a small protein with a busy agenda.Mol. Aspects Med. 2010, 31:350–358.
37.Andresen V, Pise-Masison CA,
Sinha-Datta U, Bellon M, Valeri V, Washington Parks R, Cecchinato V, Fukumoto R, Nicot
C, Franchini G: Suppression of HTLV-1 replication by Tax-mediated
rerouting of the p13 viral protein to nuclear speckles.Blood 2011, 118:1549–1559. ● This study demonstrates a new function of HTLV-1 p13 protein: when p13
and Tax are co-expressed, this latter mediates p13 ubiquitination, stabilizing
the protein which is then partially re-localized to the nucleus, where p13 binds
to Tax, preventing its association with CBP/p300 and thus reducing HTLV-1
expression.
38.Silic-Benussi M, Cavallari I, Vajente N, Vidali S, Chieco-Bianchi L, Di Lisa F, Saggioro D, D'Agostino DM, Ciminale V: Redox regulation of T-cell turnover by the p13 protein of human T-cell leukemia virus type1: distinct effects in primary versus transformed cells.Blood 2010, 116:54–62.
39.Biasiotto R, Aguiari P, Rizzuto R,
Pinton P, D'Agostino DM, Ciminale V: The p13 protein of human T
cell leukemia virus type 1 (HTLV-1) modulates mitochondrial membrane potential and
calcium uptake.Biochim. Biophys. Acta 2010, 1797:945–951. ● This paper confirms the effects of p13 on mitochondrial inner membrane
potential (Δψ) stressed previously in vitro, in living cells. Indeed, their
results showed that in HeLa cells p13, by affecting in a dose-dependent manner
the Δψ, decreases mitochondrial Ca2+ uptake.
40.Pique C, Ureta-Vidal A, Gessain A, Chancerel B, Gout O, Tamouza R, Agis F, Dokhélar MC: Evidence for the chronic in vivo production of human T cell leukemia virus type I Rof and Tof proteins from cytotoxic T lymphocytes directed against viral peptides.J. Exp. Med. 2000, 191:567–572.
41.Malim MH, Emerman M: HIV-1 accessory proteins-ensuring viral survival in a hostile environment.Cell Host Microbe 2008, 3:388– 398.


