Diploidization of meiosis in autotetraploids
2012/10/01
Abstract
Recent genome analyses indicate that genome doubling occurs frequently in eukaryotes. The presence of additional sets of chromosomes can lead to meiotic failures, which pressure polyploids to evolve into a diploid-like state. This process is called diploidization and can be broken down into two distinct types: cytological diploidization, a transition to bivalent chromosome configurations at meiosis, and genetic diploidization, the development of pairing preferences that result in disomic inheritance. In this review, I select studies that have consolidated our understanding of diploidization. Nevertheless, little is known on this subject and only assumptions can be made.
Table of Contents
- Introduction
- Autotetraploids
- Genetic diploidization
- Disomic and tetrasomic inheritance
- Meiotic pairing configurations
- Cytological diploidization observation
- Common mechanisms of cytological diploidization in allotetraploids and autotetraploids
- Some aspects that influence cytological diploidization
- Some explanations for cytological diploidization
- Conclusion
- Acknowledgments
- References and recommended reading
Introduction
Whole genome duplication, or polyploidy, is an important evolutionary force among eukaryotes. In plants, it is thought to contribute to adaptation to climate and/or edaphic changes, by increasing effective population size and decreasing inbreeding depression in the short term [1,2]. In metazoans, polyploidy is far less common, but is also believed to have led to some critical innovations in many taxa, including vertebrates [3,4]. Despite the benefits a polyploid organism may have, the process of polyploidization induces a genomic shock, leading to some evolutionary disadvantages that reduce fitness, such as genomic instability, meiotic irregularities and reduced fertility [5]. Therefore, natural selection should favor a shift back to diploid-like meiosis and genome regulation to stabilize polyploid lineages, through a process known as “diploidization”.
Polyploids are of two basic types. Those that arise from a single species are called autopolyploids, whereas those that combine the genomes of two or more related species are called allopolyploids. More is known on the mechanisms of diploidization in allopolyploids, as they are extensively studied. Despite their importance, little is known about the mechanisms of diploidization in autotetraploids; except for the recent study by Hollister et al. [6], there has only been speculation.
Autotetraploids constitute a small, but important part of the flora and fauna that we observe. There is an increasing awareness of their presence throughout the tree of life [7]. Evolutionary biology is interested in comprehending the diversity of life around us; thus it is important to better understand how autotetraploids arise and establish as successful species. Genetic diploidization is a crucial part of stabilizing autotetraploid races, so deciphering this process is key to knowing how stable autotetraploid populations are created. Since autotetraploids oftentimes have different ecological niches from their diploid progenitors [8], knowing how they arise and establish enhances our appreciation of the biological processes that create evolutionary novelty. Furthermore, challenging meiosis with modified sets of chromosomes can aid in our understanding of chromosome pairing and synapsis in general.
There are two types of “diploidization” in autotetraploids: cytological diploidization and genetic diploidization. In a cytologically diploidized organism, only bivalents are formed during meiosis, even though there are sets of four completely homologous chromosomes. In many autopolyploids, the chromosomes show no preference for pairing partner. These species have “tetrasomic” inheritance which means that each locus may independently segregate up to four alleles. Genetic diploidization can arise simultaneously or after cytological diploidization and refers to the evolution of pairing preferences for homologs over homeologs. In this case, up to two alleles may segregate at each of the two duplicated loci.
This review article will mainly focus on what is known about cytological diploidization, although genetic diploidization is also discussed.
Autotetraploids
Autotetraploids arise by genome doubling of a single diploid species through three known mechanisms (Figure 1). The most common one is through fusion of unreduced gametes. Autotetraploidy can also occur via cross-fertilization between an unreduced gamete and a diploid gamete from a triploid intermediate, also known as a “triploid bridge” [1]. For research purposes, autotetraploids can also be induced artificially by chemical induction. Colchicine, an alkaloid extracted from Colchicum autumnale and other species of Colchicum, is the most common chemical inducing polyploidy used in research. Indeed, it prevents chromosome segregation during meiosis, by inhibiting microtubule polymerization [9].
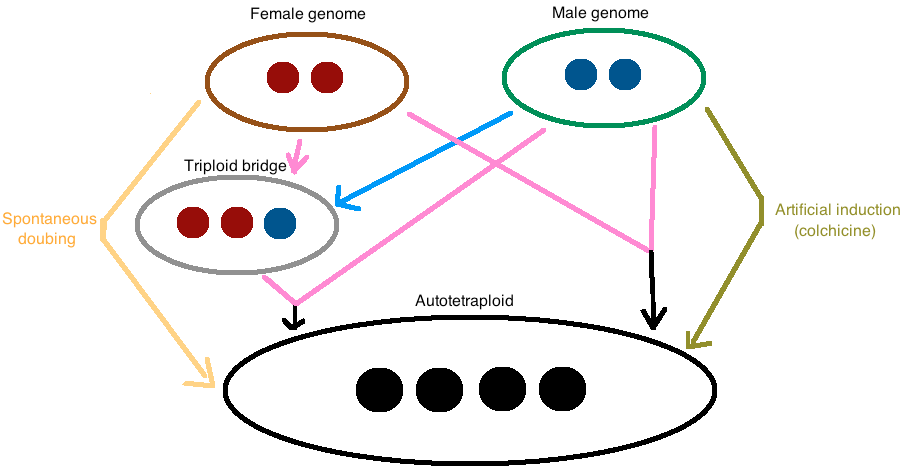
Figure 1. The major mechanisms of autotetraploid formation are shown in this figure..
Direct fusion of two unreduced gametes, cross-fertilization between an unreduced gamete and a diploid gamete from a triploid intermediate (“triploid bridge”) and spontaneous doubling. Moreover, it is possible to artificially induce autotetraploidy with colchicine. The pink arrows indicate unreduced or diploid gametes whereas the blue arrow refers to an haploid gamete.
Genetic diploidization
Genetic diploidization following whole-genome duplications in plant species as well as in animal species may have occurred quite frequently during organismal evolution [4,10]. Nevertheless, the mechanisms that lead to genetic diploidization are puzzling. One hypothesis is that chromosomal rearrangements through processes such as neofunctionalization, subfunctionalization or loss of duplicated segments, recombinations, transposable elements and genetic drift, cause differences between formerly homologous chromosomes [11]. Chromosomal changes cause them to develop pairing and synapsis preferences: among the set of homologous chromosomes, some pair with a greater probability to one particular partner – generally the most similar chromosome.
Disomic and tetrasomic inheritance
In autotetraploids, the four copies of each chromosome are derived from the same species and are therefore all homologous. This means that there is no pairing preference and any chromosome can pair with equal probability with each of the three other homologous chromosomes. This leads to a type of inheritance called polysomic, or in the case of tetraploids, tetrasomic. When pairing preferences are lacking, a chromosome can pair with different homologs at different pairing initiation sites. If these pairing interactions mature to crossovers, an association of multiple homologous chromosomes, called a multivalent, can occur during meiosis I. This can lead to problems during meiosis as these multivalent configurations may mis-segregate, resulting in aneuploid gamete formation.
However, in “extreme” allotetraploids, two sets of chromosomes are from one species and the remaining two sets are from another species. Therefore, pairing occurs between chromosomes from the same parental species (homologous) rather than between chromosomes from separate species (homeologous). This leads to a diploid-like inheritance called disomic where only bivalents form during meiosis I.
In reality, most existing autotetraploids show both tetrasomic inheritance for some chromosomes and disomic inheritance for others [12]. These complex patterns of inheritance are directly linked to meiotic configurations that are characteristic of diploidization. Therefore, in order to study genetic diploidization of meiosis, it is necessary to focus on patterns of inheritance.
Meiotic pairing configurations
In polyploids, pairing partner switches (PPS) can occur during synapsis, which means that during prophase I one chromosome can synapse with different homologous partners in different regions. This leads to the formation of a multivalent. In autotetraploids, there are two types of multivalents: quadrivalents which are composed of four homologous chromosomes, and trivalents, which are composed of three. If they are not resolved, multivalents can persist through metaphase I, leading to high rates of aneuploid gamete formation and sterility [14].
In order to understand how multivalents form, the concept of the autonomous pairing site (APS) is to be introduced. APS is the smallest chromosome region that is capable of initiating pairing between two homologous chromosomes and can hence generate a PPS, regardless of the likelihood of such PPS [15]. In the absence of any pairing preferences among homologous chromosomes and no dependence between APSs, there is a simple relationship between the number of PPSs and APSs: PPS=2/3(APS-1) [16]. The simplest model that describes multivalent formation is called the “random-end pairing model”. According to this model, pairing initiation can only occur in the distal regions of the chromosomes and each chromosome can pair with an homologous chromosome at each end with the same probability. Therefore, the multivalent frequency in prophase I is equal to 2/3, twice as much as the frequency of bivalents [17].
Comparing observed multivalent frequency to this model gives information on how cytologically diploidized an individual is but also on the number of APSs: (1) If there is only one pairing site on a given chromosome, then it can only pair with one chromosome and only bivalents are formed. Therefore, cytological diploidization is achieved for this chromosome immediately after the polyploidization event. (2) If there are two pairing sites, the number of multivalents is indicative of diploidization. Indeed if the multivalent:bivalent ratio is lower than the 2:1 ratio expected on the random-end pairing model, it can only mean that cytological diploidization is in progress. However, if this ratio exceeds the 2:1 ratio, than the chromosome must have more than two APSs.
Importantly, even if only bivalents form at metaphase I, it does not mean that there is a disomic inheritance. It is possible to have an organism with only bivalents and tetrasomic inheritances if bivalents consist of randomly chosen pairs of chromosomes. In other words, cytological diploidization can be complete while genetic diploidization is absent. The converse is, of course, not true [14].
Cytological diploidization observation
It is possible to observe the meiotic configurations in prophase I and metaphase I by electron microscopy analysis. The use of fluorescence in situ hybridization (FISH) (Figure 2) or of ammoniacal silver staining (Figure 3) as cytogenetic techniques for chromosome identification are well adapted to analyze multivalents and bivalents. However this method can be misleading since overlapped bivalents can resemble multivalents [18].
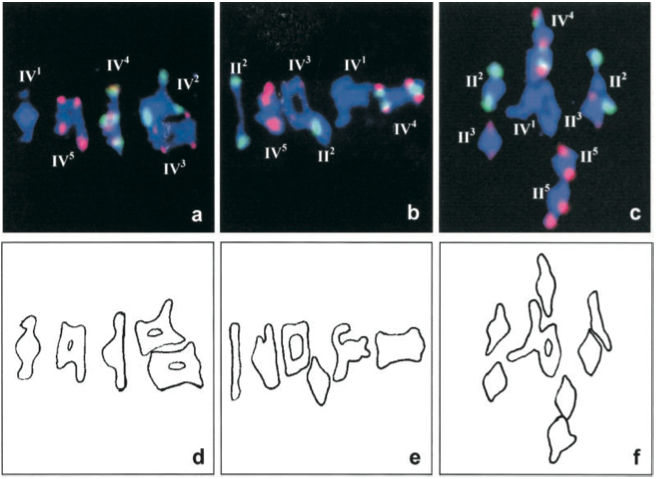
Figure 2. FISH with 5S (red) and 45S (green) rDNA probes was used to identify the different chromosomes at metaphase I of meiosis..
These three pictures show quadrivalent (IV) and bivalent (II) configurations (Reproduced from Santos et al. [14]).
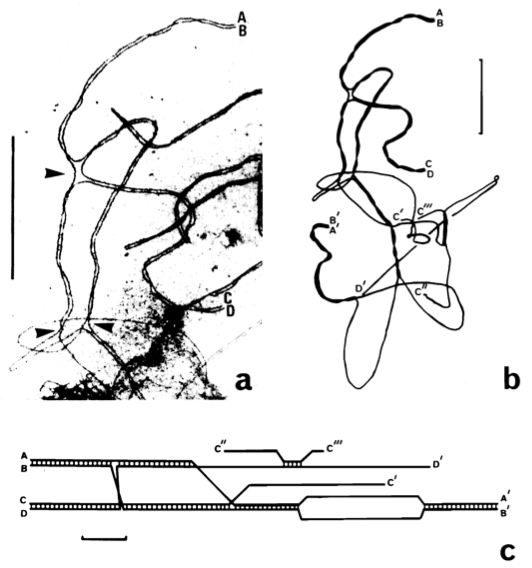
Figure 3. Structure of a quadrivalent. .
a. Electron micrograph of a portion of a quadravalent. It shows paired telomeres (AB and CD) and switch points (arrowheads). b. Tracing of the entire quadrivalent with one other paired telomere (A'B'), unpaired telomere (D'), and broken lateral elements (C', C'' and C'''). c. Diagram of this quadrivalent. Synaptonemal complex formation is indicated by cross bars between lateral elements. Bar=5µm. (Reproduced from Gillies et al. [19]).
Common mechanisms of cytological diploidization in allotetraploids and autotetraploids
Avivi [20] discovered that cytological diploidization in allopolyploids and in autopolyploids might have some common gene regulations. She defined three types of genotypes in diploid species of the wheat group, depending on the efficacy of their genes to promote or suppress pairing of homeologous chromosomes in interspecific hybrids: high-, intermediate-, or low- homeologous pairing genotypes. She noticed that in T. Longissium neoautotetraploid “low homeologous pairing” lines, there is a tendency towards bivalent pairing, which is not observed in higher homeologous pairing lines of the same diploid species. Therefore, some alleles already present in diploids regulate diploidization in wheat, and stabilize meiosis in autotetraploid derivatives.
Another similarity between allopolyploids and autopolyploids was recently discovered by Lukaszewski et al. (2010) [21]. They inserted the Ph1 locus from common wheat (hexaploid), which is known to suppress pairing between homeologous chromosomes, in autotetraploid rye and noticed that it drastically increased the rate of genetic and therefore cytological diploidization. This means that there is possibly some overlap in mechanisms that can cause diploidization of auto- and allotetraploids.
Some aspects that influence cytological diploidization
The rate of cytological diploidization varies among species and accessions. It is also affected by the size of chromosomes and the ploidy level.
Species dependance:
Curole et al. (2005) [12] surprisingly found not only cytological diploidization, but genetic diploidization in second-generation autotetraploid Pacific oysters, initially produced by chemical treatment. Indeed, the high levels of preferential pairing in the second-generation autotetraploid Pacific oysters are indicative of genetic diploidization and a fortiori of cytological diploidization. For that matter, this example also suggests that structural differences or genetic divergence emerged within the previous two generations or were present in the founders of these tetraploid lines.
An additional example from the animal kingdom is the salmonidae family. Though it's hypothesized that a single autotetraploid ancestor gave rise to all the species in this family, some of these species are completely genetically diploidized whereas others are only partially. The process of diploidization seems to be in progress even though the polyploidization event probably occurred about thirty million years ago [22].
Furthermore, even within a single species, there can be differences in diploidization rate between lines that have existed at the tetraploid level for the same amount of time [14,20]. This promotes the idea that stochasticity might play a role in diploidization rate as well.
Size dependance:
Santos (2003) [14] noticed that in established Arabidopsis thaliana autoteraploid lines, the shorter chromosomes (2 and 4) exhibit lower multivalent frequencies than the longer ones. He concluded that these lines have undergone a partial cytological diploidization of meiosis. G. H. Jones (1994) [16] noticed that in Crepis capillaris PPS frequency increases almost linearly with chromosome size, which leads to the same conclusion. The best explanation for this is that the number of autonomous pairing sites and chiasma are lower in smaller chromosomes.
However, this is not true interspecifically. For example, Crepis capillaris and Triticum monococcum are two species that have similar chromosome sizes but have very different multivalent frequencies [23], while colchicine-induced Arabidopsis thaliana autotetraploids have similar multivalent frequencies to Crepis capillaris but have ten times smaller chromosomes [14].
Ploidy level dependance:
The number of chromosomes within a same species, that is to say the ploidy level, also seems to affect cytologicaI diploidization. In fact, Hamey et al. (1988) [15] showed that within each chromosome group in Crepis rubra, there are more multivalents in the autotriploid lines than in the autotetraploid lines, although it contradicts theoretical considerations [16]. The same phenomenon was observed in Crepis capillaris [16].
However, it isn't mentioned which lines of Crepis rubra were used for this study. Moreover, for his study, Jones used established lines of Crepis capillaris in which the polyploidization events' origins are unknown. Therefore, we can't be sure that the differences between autotriploids and autotetraploids are not due to the fact that the autotetraploid lines are older than the autotriploid lines and accordingly, “had more time” to diploidize. Avoiding this problem could be done by inducing triploidy and tetraploidy artificially and to observe, in parallel, how both lines evolve. If this experiment leads to the same conclusion, this result could be correlated to that of Bennet and Smith who showed, without being able to explain why, that the duration of meiosis decreases as the ploidy level increases for related species, at least in some organisms [25]. And so, one explanation is that multivalents have less time to form in autotetraploids than in autotriploids.
Another explanation is that when an odd number of chromosomes is present, competition between homologous chromosomes arises for pairing partners. In autotriploids for example, if two chromosomes start to pair, the unpaired third chromosome will be actively seeking to pair at any of the APSs it can find, which can lead to aneuploidy or other meiotic failures [16].
Some explanations for cytological diploidization
There are a few explanations for cytological diploidization that can be found in literature. None of them is completely verified, but all seem plausible and are not mutually exclusive.
Sved (1966) [25] hypothesized that a low quadrivalent frequency can be explained if strands fail to initiate pairings or if there is insufficient chiasmata formation to maintain all pairings. This seems to be the case of Dactylis glomerata according to McCollum study (1958) [26]. However, this isn't the case of T. Longissium, where the chiasma frequency and the number of paired chromosomal arms are the same in more diploidized lines and in less diploidized lines. Thus, though there are suggestive correlations, pairing initiation cannot alone explain all observed patterns.
In the same year, Feldman (1966) [27] published his “spatial hypothesis” according to which the meiotic behavior of homologous chromosomes is predetermined by their spatial configurations in somatic and premeiotic cells. The frequency of multivalents would be directly linked to how associated the homologous chromosomes are to each other before meiosis begins. However, this hypothesis is controversial. On one hand, Vega and Feldman (1998) [28] support it with some experimental evidence, but on the other hand, it contradicts the idea that all premeiotic contacts are lost during DNA replication [29].
Another possibility is the interference between two APSs from a same chromosome. As a matter of fact, the pairing partner choice at an APS may be physically disturbed by the pairings that occur at adjacent APSs. If two APSs are close enough, it's more likely that the partner will be the same for both of them, without there necessarily being pairing preferences. It is thus decreasing the frequencies of PPSs and as a consequence increasing cytological diploidization (by increasing bivalent formation). This type of interference was named “negative pairing interference” by G. H. Jones (1994) [16]. He also defined another type of interference, a “positive pairing interference”, in which two adjacent APSs would rather pair to different chromosomes than to the same one.
Very recently, Hollister et al. [6] compared the genomes of cytologically diploidized autotetraploid Arabidopsis arenosa individuals to diploid Arabidopsis genomes. They found 192 genes with evidence of recent selective sweeps of which several are involved in stabilization of meiosis. These genes encode proteins implicated in synapsis, chromosome cohesion and recombination. A special emphasis was put on the crossover related gene ASYNAPSIS1 (ASY1). This gene is of particular interest because it is a target of Ph1, the only cloned diploidization gene. A radical amino acid substitution in a highly conserved domain of the protein was found to be common in autotetraploid Arabidopsis arenosa and rare in the diploids. This gives reason to ask whether this allele has any effect on crossover rates in either genetic context. Therefore, this study is promising when it comes to unraveling some important genetic factors for diploidization.
Conclusion
Despite the evidence of polyploidization as an important event in the evolution of many plant and animal taxa, the molecular bases of cytological diploidization as well as of genetic diploidization remain mysterious. Some hypotheses have been suggested but need to be elaborated. They include pairing interferences, pairing initiation failure, pairing preferences, low level of chiasmata formation, premeiotic configurations and genetic factors. Moreover, the rate of diploidization seems to depend on various aspects such as chromosome size and ploidy level and varies among species and accessions (Figure 4).
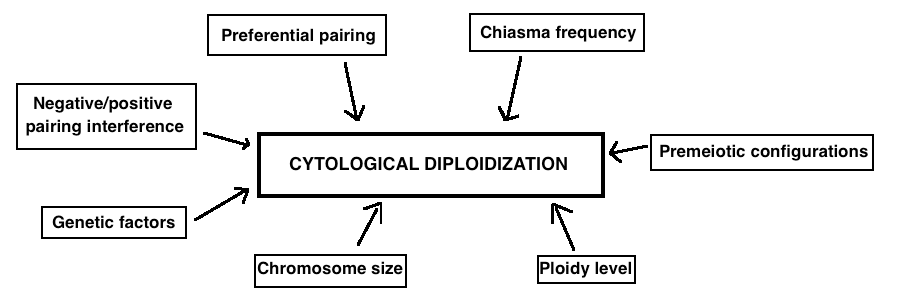
Figure 4. The main factors that are believed to influence cytological diploidization are listed in this figure..
It is important to bear in mind that there is not enough evidence to prove or to contradict any of these assumptions. They all need to be challenged.
A first step to understanding diploidization should be to distinguish the processes of cytological diploidization and genetic diploidization. They tend to be confused with each other because genetic diploidization is a specific type of cytological diploidization that results from preferential pairings, but mostly because in allopolyploids, they usually come hand in hand. This is probably rare in autopolyploids where cytological diploidization can occur with no evidence of any preferential pairing whatsoever.
Acknowledgments
The author gratefully acknowledges the contributions of Professor Kirsten Bomblies, Dr. Ben Hunter, Brian Arnold and Emma Sedivy for editorial suggestions.
References and recommended reading
Papers of particular interest have been highlighted as:
● of special interest
●● of outstanding interest
1.Parisod C, Holderegger R, Brochmann C: Evolutionary consequences of autopolyploidy. New Phytol 2010, 186:5-7.
2.Brochmann C, Brysting A.K, Alsos IG, Borgen L, Grundt HH, Scheen A-C, Elven R: Polyploidy in arctic plants. Biol J Linn Soc 2004,82:521–536.
3.Furlong RF, Holland PWH: Polyploidy in vertebrate ancestry: Ohno and beyond. Biol J Linn Soc 2004, 82:425-430.
4.Ohno S:Evolution by Gene Duplication. Springer-Verlag; 1970.
5.Ramsey J, Schemske DW: Pathways, mechanisms and rates of polyploid formation in flowering plants. Annu Rev Ecol. Syst. 1998, 29:477-501.
6.Hollister JD, Arnold B, Svedin E,
Xue K, Dilkes B, Bomblies K: Genetic adaptation associated with
genome-doubling in autotetraploid Arabidopsis
arenosa.PloS Genet. 2012. ●● In this study the authors provide candidate genes and mechanisms to
further our understanding of the molecular mechanisms underlying autotetraploid
stabilization and set the stage for developing Arabidopsis
arenosa as a genetic model for understanding autotetraploid
evolution in molecular detail.
7.Soltis DE, Soltis PS: Polyploidy: Recurrent formation and genome evolution. Trends Ecol Evol 1999 14:348–352.
8.Ramsey J: Polyploidy and ecological adaptation in wild yarrow. Proc Natl Acad Sci USA 2011, 108:7096–7101.
9.Kokate, Jalalpure, Hurakadle: Textbook of Pharmaceutical Biotechnology, p.80. Elsevier; 2011.
10.Mitchell-Olds T, Clauss MJ: Plant evolutionary genomics.Curr Opin Plant Biol 2002, 5:74-79.
11.Ohta T: Evolution of gene families.Gene 2001, 259:42-52.
12.Curole JP, Hedgecock D: Estimation of Preferential Pairing Rates in Second-Generation Autotetraploid Pacific Oysters (Crassostrea gigas).Genetics 2005, 171:855-859
13.Loidl J: Cytological aspects of meiotic recombination. Experientia 1994 50:285-294.
14.Santos JL, Alfaro D,
Sanchez-Moran E, Armstrong SJ, Franklin FCH, Jones GH: Partial
Diploidization of Meiosis in Autotetraploid Arabidopsis
thaliana.Genetics 2003, 165:1533-1540. ●● By analyzing the different chromosomes of Arabidopsis
thaliana with fluorescent probes, the authors concluded that the
studied lines have undergone a partial diploidization of meiosis. Moreover, the
reduction in multivalents was higher in the smallest chromosomes.
15.Hamey Y, Abberton MT, Wallace AJ, Callow RS: Pairing autonomy and chromosome size. In: Brandham PE (Ed) Kew Chromosome Conference III, p.241–251. Her Majesty's Stationery Office; 1988.
16.Jones GH: Meiosis in autopolyploid Crepis capillaris. III. Comparison of
triploids and tetraploids; evidence for non independence of autonomous pairing
sites.Heredity 1994 73:215-219. ● In Crepis cappilaris, cytological diploidization is
affected by the ploidy level. Indeed, chromosomes of autotriploids form 60% more
PPSs than autotetraploids.
17.Jackson RC, Casey Y J: Cytogenetic analyses of autopolyploids: models and methods for triploids to octoploids. Am J Bot 1982, 69:486-501
18.Weiss H, Maluszynska J: Chromosomal rearrangement in autotetraploid plants of Arabidopsis thaliana.Hereditas 2000, 133:255-261.
19.Gillies CB, Kuspira J, Bhambhani RN: Genetic and cytogenetic analyses of the A genome of Triticum monococcum. IV. Synaptonemal complex formation in autotetraploids.Genome 1987, 29:309-318.
20.Avivi L: The
effect of genes controlling different degrees of homoeologous pairing on
quadrivalent frequency in induced autotetraploid lines of Triticum
longissimum.Can J Genet Cytol 1976, 18:357-364 ● A clear demonstration that some genes that control meiotic pairing and
diploidization do not arise de novo in response to
polyploidization but are present in diploid species.
21.The Ph1
locus from wheat controls meiotic chromosome pairing in autotetraploid rye
(Secale cereale L.).Cytogenet Genome Res 2010, 129:117-123 ●● This paper demonstrated that the Ph1 locus from
common wheat dramatically accelerated the process of diploidization in
autotetraploid rye. This fascinating fact suggests that some aspects of
diploidization are universal.
22.Stouder DJ, Bisson PA, Naiman RJ:Pacific Salmon And Their Ecosystems, p31. Chapman & Hall; 1996.
23.Jones GH, Vincent JE: Meiosis in autopolyploid Crepis capillaris. II. Autotetraploids.Genome 1994, 37:497-505.
24.Bennett MD, Smith JB: The effects of polyploidy on meiotic duration and pollen development in cereal anther. Proc R Soc Lond B 1972, 181:81-107
25.Sved JA: Telomere attachment of chromosomes. Some genetical and cytological consequences.Genetics 1966, 53:747-756.
26.McCollum GD: Comparative studies of chromosome pairing in natural and induced tetraploid Dactylis.Chromosoma 1958, 9:571-605
27.Feldman M: The effect of chromosomes 5B, 5D and 5A on chromosomal pairing in Triticum aestivum.Proc. Natl. Acad. Sci. USA 1966, 55:1447-1453.
28.Vega JM, Feldman M: Effect of the Pairing Gene Ph1 and Premeiotic Colchicine Treatment on Intra- and Interchromosome Pairing of Isochromosomes in Common Wheat.Genetics 1998, 150:1199-1208.
29.Zickler D: From early homologue recognition to synaptonemal complex formation.
Chromosoma 2006,
115:158-174. ● A thorough review on fundamental aspects of chromosome pairing and
synapsis.


