Morphogenesis of the flower of Arabidopsis,genes networks and mathematical modelling.
2012/10/01
Abstract
Organ formation during flower development in Arabidopsis is regulated by only few genes, what can be summarized by the so-called 'ABC' model. Interactions between these genes and with other non-ABC genes are necessary to establish the final pattern of concentric circles of ABC gene expression. To understand this process, the expression and the interactions between these genes in the early stages of development are investigated experimentally. These observations allow us to build discrete and continuous models which can allow to understand pattern formation and stability mechanisms and help to design new experiments.
Table of Contents
Introduction
Flower are present in a great number of different plants in nature, including over 200,000 species of dicotyledonous plants. With very different geometries they generally have the same role in plant reproduction. The development of the flower was well conserved during the evolution of dicotyledons and is rather the same between different species. The most common model system to study the flower development is Arabidopsis Thaliana. Its development is well described and is divided into several 'developmental stages', from floral to seed dispersal [1]. Here, I will review the literature pertaining to the first few stages, up to arrive to the establishment of the ABC pattern as well as those that pertain to model flower formation
1. Flower initiation
The floral meristem emerges as a lateral outgrowth from the periphery of the inflorescence meristem. Only a few genes are necessary to confer floral meristem identity. LEAFY (LFY) and APETALA 1 (AP1) are genes that ensure that primordia adopt a flower fate [2]. These two genes are expressed very early and are in mutual repression with TERMINAL FLOWER 1 (TFL1), a gene that maintains the inflorescence meristem's identity. In parallel, AP1 is regulated by at least two independent pathways. LFY is part of one of them in regulating positively AP1 that in turn can positively regulate LFY expression [3,4]. The flower initiation results from oriented cell division and growth. Models proposed by Kwiatkowska explain the role of these factors in the initiation process. These models give tools to represent meristem or others plant tissues. [5,6].
2. The ABCE system
The flower is composed of four organ types separated in four different concentric circles called whorls. In the wild type flower of Arabidopsis, four sepals can be observed in the outer whorl, which form with the four petals in the second whorl the perianth. Then one finds six stamens which produce the pollen, and in the fourth whorl, the gynoecium, composed of two carpels which produce ovules. To lead to this organ formation, Smyth has separated the flower formation in a few stages [1] : Stage 1 is the formation of a bump at the periphery of the inflorescence meristem, stage 2 begin at when a border appears between the meristem and the developing flower. At stage 3, sepal formation can be observed and at stage 6 the future organ can be distinguished. The four whorls that predict the organ position are characterized from the stage 3 of the flower development by the expression of three classes of genes (Figure 1.2).The function of these different genes can be understood by observing the morphology of different mutants : ap1 and ap2 give carpels instead of sepals and stamens instead of petals ; ap3 and pi give sepals instead of petals and carpels instead of stamens and finally ag gives petals instead of stamens and sepals instead of carpels. These observations explain the ABC system (Figure 1.1) with class A genes, APETALA 1 and 2 (AP1 and AP2), present in sepals and petals, class B genes, APETALA 3 (AP3) and PISTILLATA (PI), present in petals and stamens and class C genes, AGAMOUS (AG) present in stamens and carpels [7]. Except for AP2, all these genes code for transcription factors which play a key role in organ initiation.
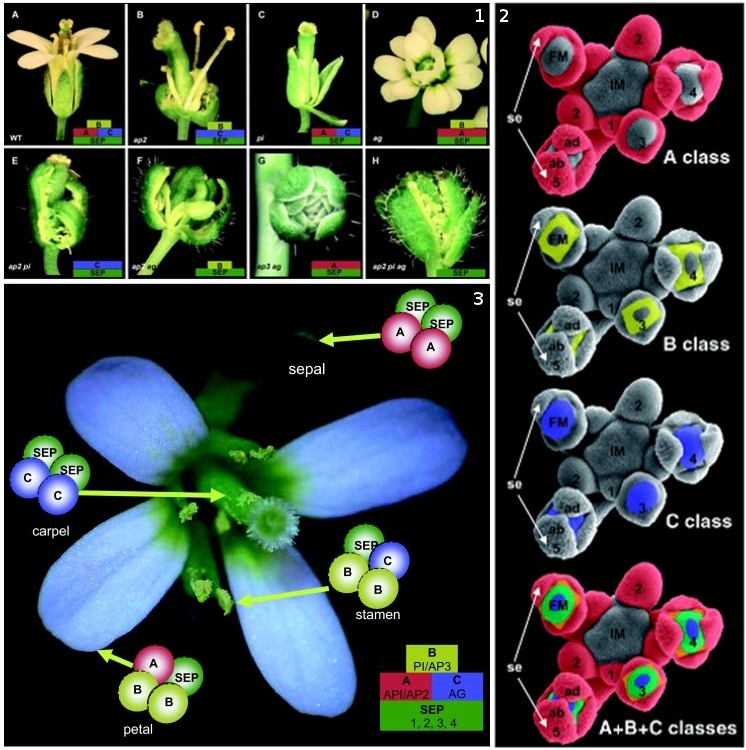
Figure 1. ABC model of floral-organ patterning. .
Arabidopsis homeotic floral mutants which explain the ABC model based on experiments from Coen [7]. 1.A. Wild-type flower. 1.B. Single ap2 mutant composed of carpelloid sepals, stamens, stamens and carpels. 1.C. Single pi mutant composed of sepals, sepals, carpels and carpels. 1.D. Single ag mutant has the stamens transformed into petals and the carpels are replaced by another flower repeating the same pattern. 1.E ap2,pi double mutant composed only of sepalloid carpels. 1.F. ap2,ag double mutant has leaf-like organs in the first and fourth whorls and mosaic petal/stamen organs in the second and third whorls. 1.G.ap3,ag double mutant with repeated whorls of sepals. 1.H. ap2,pi,ag triple mutant which has leaf-like organs with some residual carpel properties. 2. Expression of the A,B and C genes during the early stages of the flower development. 3. Representation of the quartet model in the wild-type flower [8]. (Reproduced from [8]).
2.1. Class A genes
AP1 and AP2 form the class A genes. AP1 is expressed everywhere in the floral meristem at the early stages of development and disappears from the two inner whorls during stage 3 (Figure 2). This is due to repression by AG [9]. AP2 is also expressed everywhere in the floral meristem and play a role in its identity [3], but its activity is repressed by MicroRNA miR172 in the two inner whorls. AP2 is also repressed by AG in this area [10,11].
2.2. Class B genes
PI and AP3 are the two genes of the class B, there are expressed in the second and the third whorls from stage 3 (Figure 2). They can auto-regulate themselves by mutual activation and by self-regulation. Nevertheless, a small difference between this expression is found. PI is expressed at the early stage 3 in the center of the floral meristem and disappears then from the inner whorl, in contrast to AP3 which is never expressed in this whorl [12,13]. AP3 and PI proteins can form an heterodimer. This heterodimer can also bind to the promoter region of AP1 and repress AP1 [14]. PI and AP3 are also positively regulated by the class A and C genes [15].
2.3. Class C gene
AG is the only known class C gene. It is expressed in the two inner whorls from stage 3 (Figure 2). AG represses expression of the class A genes AP1 and AP2 [16-18]. Class A and class C genes are antagonists. This is confirmed by observations of expression of AP1 everywhere in ag mutant and of AG everywhere in ap1 and ap2. AG interacts also with AP3, PI and SEPALLATA 3 (SEP3), which are together regulated by a positive-feedback loop to maintain their own expression [19].
2.4. The quartet model
There are a fourth class of genes, the class E genes which are the four SEPALLATA genes (SEP1-4). These genes have redundant activities and are necessary for ABC genes activity and organ formation. The transcription factors AP1, AP3, PI, AG and SEP are MADS domain proteins. They can form dimers to bind to their CArG (CC(A/T)6GG) box DNA target sequence. Four tetramers are present in the four whorls, they predict organ identity: AP1/SEP/AP1/SEP specify sepals, AP1/SEP/AP3/PI petals, AG/SEP/AP3/PI stamens and AG/SEP/AG/SEPcarpels. The model of these tetramers formation is called 'the quartet model' (Figure 1.3). These quartets are formed thanks to DNA looping and the presence of SEP proteins, which help to form tetramers [20‑22]. The SEP1 and 2 genes are expressed everywhere while SEP3 is expressed in the three inner whorls and SEP4 in the first (Figure 2). sep triple mutant is needed to see loss of function in the flower [23].
3. Regulation of ABC genes
The ABCE system explains well the apparition of the different organs of the flower, but is not sufficient to explain the establishment of the pattern. Some transcription factors are important in establishment, in floral identity, in earlier stages of development or in stop growth determination.
UNUSUAL FLORAL ORGANS (UFO) is one of them. It is required for the proper identity of the floral meristem. It acts very similar to LFY, and controls cell proliferation during organ growth. In the first stages of development, UFO regulates positively AP3 expression. It seems that UFO degrades proteins which repress AP3 [27]. Recent studies show that this F-box protein acts with LFY as a transcriptional co-factor to activate AP3 expression [28]. It is observed that UFO expression prevents AP3 expression. Indeed, UFO is present everywhere in stage 2, then it reduces its expression in the whorls 2 and 3 to progressively be concentrated in the second whorl at stage 6 [29].
Another pathway which regulates the ABCE pattern has LEUNIG (LUG) for principal gene. LUG in association with SEUSS (SEU) regulates AG expression [18] and also the class B genes during the two first stages of flower development. These genes need the expression of other transcription factors. AGAMOUS-LIKE 24 (AGL24) promotes the floral meristem identity and SHORT VEGETATIVE PHASE (SVP) represses floral meristem identity. These two genes are also expressed in the early stages of flower development (Figure 2). They are repressed by AP1 and LFY. AGL24 and SVP form dimers with AP1 (AP1-AGL24 and AP1-SVP), which interact with LUG-SEU dimers to repress the B and C classes genes [30].
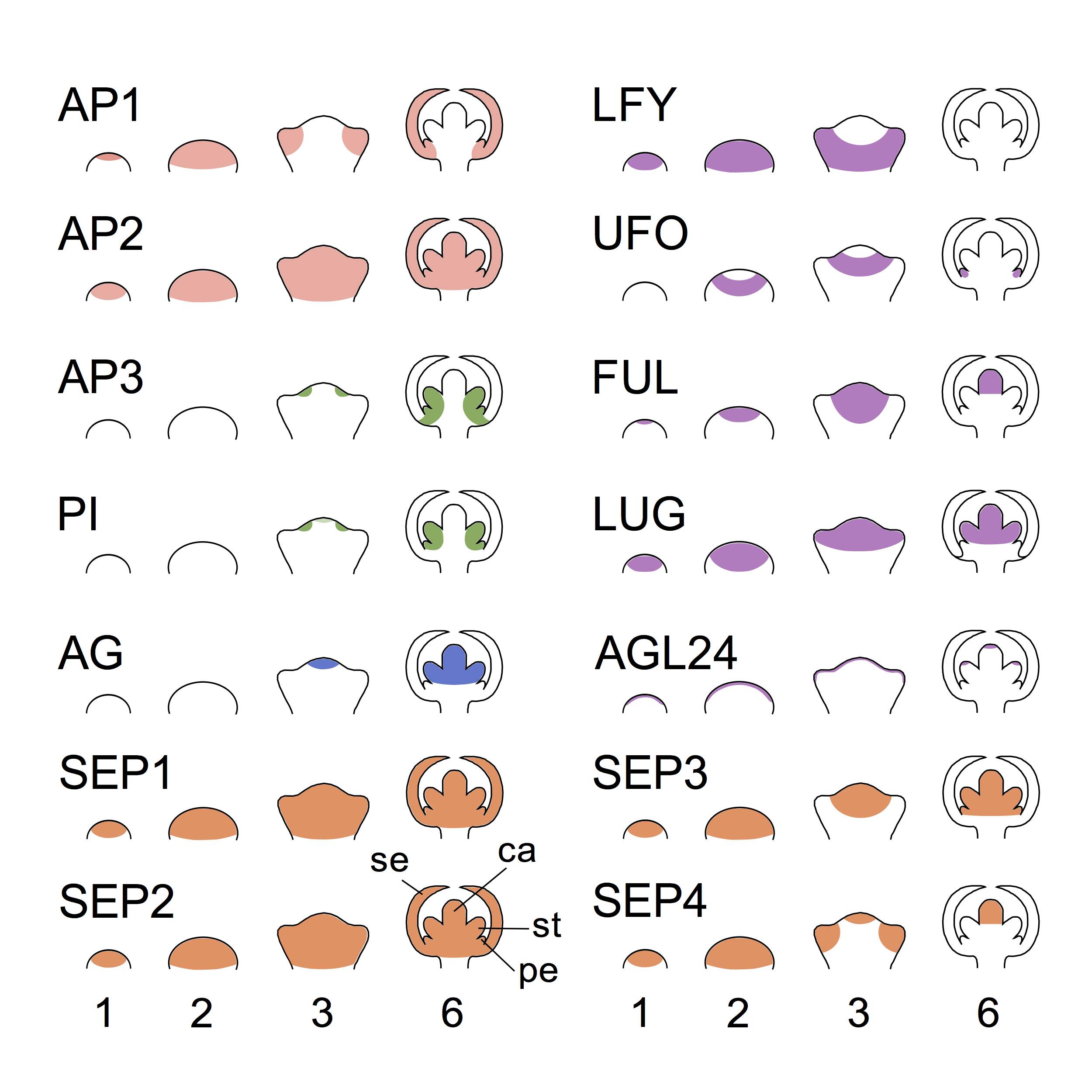
Figure 2. Pattern expression of the genes involves in the establishment of the ABC system at the earliest stages of flower development :.
Based on published gene expression data (obtained by proteins expression observations) [8,24‑26], this figure show the expression of class A (AP1, AP2 ), class B (AP3, PI), class C (AG), class E (SEP1-4) genes as well as non-ABC genes (LFY, UFO, FUL, LUG and AGL24) at stages 1, 2, 3 and 6 of the flower development. The chosen non-ABC genes play a key role in formation of the ABC pattern, which stabilizes at stage 6. Stage 1 and 2 show the bump formation. At stage 3, future sepal appears. Sepal (se), petal (pe), stamen (st) and carpel (ca) are indicated at stage 6 can be observed.
4. Modelling of pattern formation in the flower
Like we have seen in previous sections, flower development involves the expression of a certain genes and protein interactions. Finally, different patterns that depend on these interactions are obtained. To understand the existence, the stability and the establishment of this special pattern, mathematic models and numerical simulations are necessary. Few proposed model are based on observations of the different gene interactions mainly obtained by mRNA expression patterns under wild type and mutant background that allow the understanding of related pathways. With confocal microscopy and the use of computational models, it is possible to simulate the meristem development by cell identification and then to model interactions between proteins and gene expression. Recent studies show, for example, how meristem properties are maintained due to the expression of WUSCHEL (WUS), CLAVATA 3 (CLV3) and auxin which maintain the stem cells in the center of the meristem [31]. The use of deterministic ordinary differential equations (ODEs) to model different interactions, adapted in the description of the meristem geometry let to a better understanding of the system behind WUS expression [32].
4.1. Genetic Regulatory Networks
After the observation of the complexity of certain genetic pathways, Kauffman introduces in 1969 a model of Genetic Regulatory Network (GRN) by logical considerations [33]. This method is well studied by Aldana [34]. We start with a Boolean net of N elements {σ1,σ2, ...,σN}, each of them is a binary variable σi ∈ {0,1}, i = 1,2, ...,N that corresponds to the N genes expression of our system. σi = 1 if the ith gene is expressed and σi = 0 if it is not. At fixed time, each of these elements is given by a function of other elements depending on interactions between the different genes. Then the value of σi at time t + 1 is given by the value of its Ki controlling elements σj1(i)(t), σj2(i)(t), . . . , σjKi(i)(t) at time t :
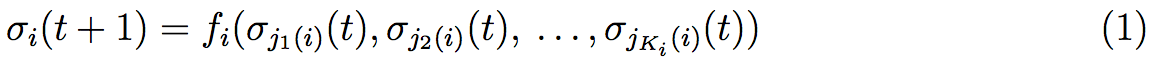
where fi is a Boolean function associated with the ith element that depends on its controlling elements. fi functions can be fixed during all the iterations. In that case the function is given by a table of interactions. Nevertheless, these functions can change during the dynamic of the system. After a few iterations with some given initial conditions, fixed points or fixed orbits that are generally called attractors can be obtained. These different attractors can be compared by their differences (we can measure these differences for example using the Hamming distance) to understand the possible transitions between the different attractors. The evolution of these differences allows to know if the system is chaotic (without stable attractors) or stable.
Generally, interactions between proteins are well known in the system, but the stoichiometry of the different reactions stays unknown. It is the principal reason of the use of discrete models, such as Boolean models. Nevertheless, Mendoza, [35], proposed a standardized methodology that renders a dynamic model of regulatory networks, where interactions are known but not necessary the biochemical reactions. The method starts with a discrete model governed by logical rules as in preceding studies. Then, the steady states of this discrete dynamical system can be easily obtained. At the same time, the network is converted into a continuous system given in the form of ordinary differential equations:
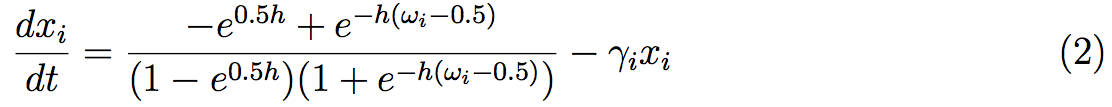
The equation describes the rate of change of the protein xi concentration. The dynamic involves activation/inhibitory effects of regulators ωi and the degradation of the protein given at the γi. This model uses the sigmoid function h that usually represent protein concentration. The study of the ODEs system uses the steady states of the discrete system. Then we can study the stability and the robustness of the steady states and obtained, thanks to variations, other steady states. The combination of the two discrete and continuous systems allows a better understanding of the regulatory network.
These theoretical approaches are applied to many biological systems, Belleza [36] applies the GRN model to four biological systems, one of them is the flower pattering in Arabidopsis. The application of such methods shows that the model should be robust to have stable attractors, but it also should be flexible to allow the creation of different attractors (think of the four whorls of the flower). Modelling needs to find a balance between order and chaos. It is the critical area studied by Aldana [34]. This convergence near the critical area can be explained by the evolution of the system near a robust and stable system that can be easily conserved between species [37].
4.2. Applications of GRN computing methods on the flower patterning
These theoretical approaches can be applied to study of the establishment of the ABC system. Summarizing the different interactions seen in the first part with some additional genes, the existence of the ABC system can be explained. A first approach consists of the study of a binary system of eleven genes (the ABC genes AP1, AP3, PI, AG and other transcription factors of the identity of the floral meristem and the activation or repression of the ABC genes: LFY, EMBRYONIC FLOWER 1 (EMF1), TERMINAL FLOWER 1 (TFL1), CAULIFLOWER (CAL), LUG, UFO and SUPERMAN (SUP)) with comparisons between the power of the different interactions. After the creation of a table of interactions and iterations of the model, the system renders six attractors, four of them correspond to the four whorls of the flower, while one corresponds to the non-floral cells and the sixth is naturally non-existent but can be easily induced by mutant experiments [38]. These six attractors can also be deduced from a reduced system of ten genes without CAL with three levels of expression 0, s and 1. This system explains that certain genes have less importances in the existence of the four flower attractors. EMF1 is a floral repressor gene only expressed in non-floral attractors, and UFO, LUG and SUP regulate the fine spatial expression patterns of organ identity genes, so they only play a role in the activation of genes like AP3, PI and AG. Thus, only TFL1, LFY, AP1, AG, AP3 and PI are sufficient to understand the existence of the four attractors [39].
More recent analysis of the fifteen genes system explains the robustness of the four attractors. This system depicted in Figure 3 with three levels of gene expression is used to explain the different patterns of mutants like ap2, lug, ap3... or 35S::AG transgenic lines. The interactions of the different mutants are resumed in tables and analysis of the logical rules gives patterns observed in experiments [15]. This system is the first that really explains the ABC system. Chaos has used again the same GRN but with only two levels of expression. He has obtained with this system the same attractors than the three levels system with a quantitatively identical manner of perturbations responses. This shows that Boolean system are sufficient to explain the existence of four attractors. He has also shown that the system has a good robustness [40].
This system of fifteen genes renders in reality ten different attractors, four are present in the inflorescence meristem, the six other in the floral meristem with two attractors for petals and two for stamens formation within or without the expression of UFO. These results are obtained using Markov chains. The four first attractors are very similar, and further to the six other attractors. It shows the importance of floral identity genes. In addition to the discrete study, another consists to use the Glass model. The Glass model adds stochastic perturbations to the boolean GRN model. The model fixes a probability η that the result is the contrary of the deterministic prediction. This probability can be between 0.5 and 10%. We obtain similar equation than in equations (1) and (2):
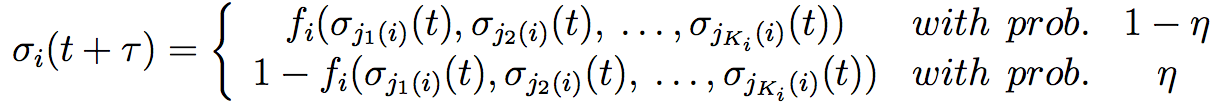
This system yields qualitatively similar results as the Boolean system but not quantitatively identical. The simulation of temporal transitions among attractors mimics the sequence of the apparition of A, B and C genes expressions. In fact, the continuous model with perturbations gives at first the apparition of the pattern configuration associated of sepals, then of petals, stamens and finally carpels. This order corresponds perfectly to the succession of the four whorls in the flower. A map of the transition gave by Alvarez-Buylla [41] explains geometrically transitions between the different attractors.
The updated system (Figure 3) gives the same attractors than previously, this confirms the robustness of the system. With a new continuous system, it is possible to obtain new steady states that can explain transitional phases during the flower pattern establishment. This continuous system is quite similar to those of Mendoza [35], but with different specific logical rules. Then, they obtain, with coupled ODEs, fourteen new transitional steady states that can be present during the establishment of the pattern [42].
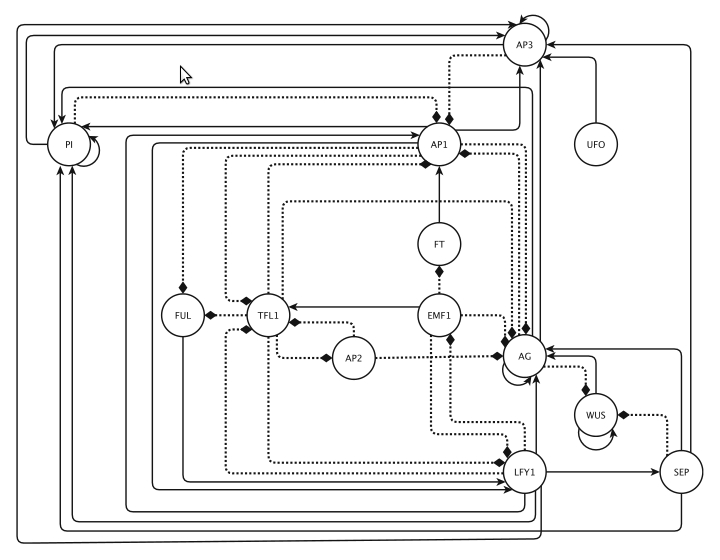
Figure 3. : Floral organ specification gene regulatory network. .
Based mainly on mRNA expression patterns. This GRN is updated from Espinosa-Soto [15]. It controls the early differentiation of inflorescences and floral organs in Arabidopsis thaliana. Positive and negative regulatory interactions are represented by continuous and discontinuous arrows respectively. (Reproduced from [42]).
4.3. Simplified models
The different models present in the previous section allow to understand the existence of different steady states found in the four different whorls and few steps to lead to these steady states. Nevertheless, these models do not explain the organization of these steady states into four concentric circles. Barrio has proposed recently a spatio-temporal model [43]. He has simplified the floral meristem to have a spherical shape, which kep constant during simulations. To understand the spatio-temporal establishment, two aspects are necessary. First, a physical field is required to break the symmetry of the spatial domain into different regions where the transcription factors can interact together. A phase-field model can be used to include this physical property. This field is an abstraction with no underlying biological mechanism. Second, a simple GRN, that is sufficient to explain the establishment of the different whorls, is associated to the phase-field model. Barrio has introduced two parameters: a 'phase-field' Φ and the 'spontaneous curvature-like' u that follow differential equations. These equations are coupled with a simple GRN of four genes: AP1, AP3, AG and WUS with the classical rules detailed in previous sections. Numerical simulations confirms the possibility of obtaining four whorls and then the system is robust to parameter variations. This model is also able to mimic patterns of several mutants.
The last model proposed to explain the establishment of the four whorls pattern was proposed by Ming [44]. This model starts with a GRN of three transcription factors (A, B and C) with auto-regulation of A and C and antagonism of these two transcription factors. B expression is activated in presence of A and C and auxin activates A and C expression. These transcription factors are modelled by three ordinary differential equations based on a function f of activation probability of the transcription of the gene X by the protein Y :
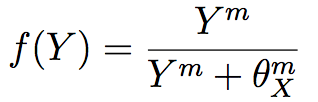
where θXm are the concentration of Y at which f = 1/2 and m is the number of Y binding sites. If Y represses X, then we use 1 − f(Y). The fourth ODE explains the transport of auxin by PIN FORMED1 (PIN1) one of the auxin efflux facilitators. These equations are integrated in a system that counts for the cell division. Then, with simulations, the model is able to capture aspects of the wild-type pattern described in previous sections. However this system stays inconsistent, possibly due to the small number of transcription factors considered [44].
These different models use some interactions between proteins to predict how proteins production will evovle in the cell. Nevertheless cell-cell communication by diffusion or transport are generally omitted. Actually, few studies explain the importance of diffusion in plant models, for example by using diffusion in the GRN system [45]. The existence of a gradient of protein concentrations can also be obtained without diffusion equations, as it was shown in a growing tissue by Chisholm [46].
5. Conclusion
The flower development (summarized in Figure 4) is a complex system of protein reactions that lead to the establishment of a four whorls pattern. There are many different proteins that play a role in this establishment. They interact together through activation or inhibition pathways. These interactions are well known now. With the use of mathematical modelling, it may be possible to determine which proteins play the key role in flower development. Most of the models can explain the existence and the robustness of the system, some even the possible establishment of whorls but actually none of them explains the early first stages that result in the ABC system creation. It is an important field of research that needs further exploration. This special system of concentric circles is also present in other living organisms like for example in the leg of the Drosophila [48], which in early phases of development exhibits various whorls. Will models of flower development be applicable to Drosophila ?
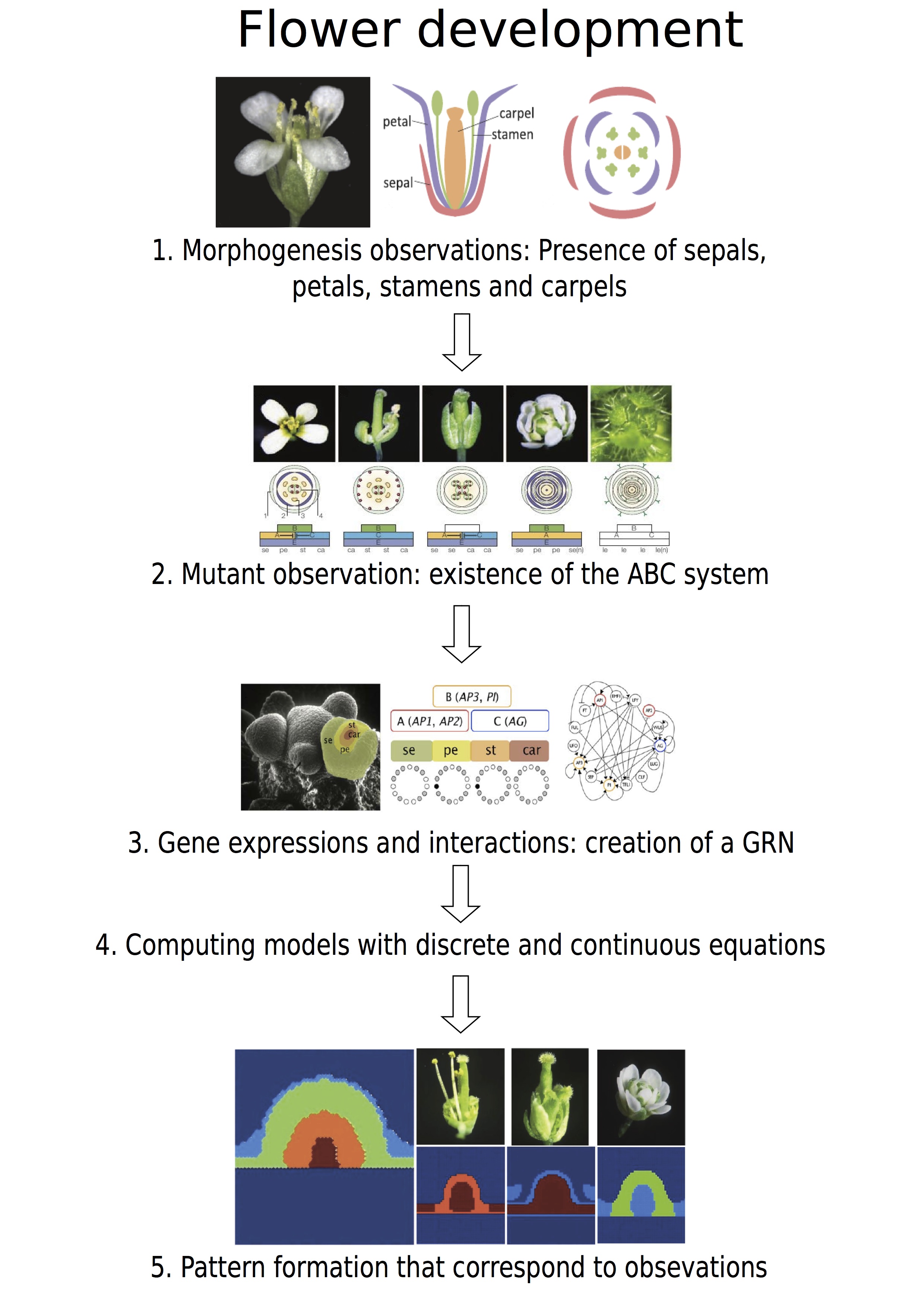
Figure 4. Resumed figure about the flower development. .
1. Observations of the flower from Irish [47], 2. Mutant observations from Krizeck [24], 3. Genetic regulatory network from Alvare-Buylla [41], 5. Simulation from Barrio [43].
References and recommended reading
Papers of particular interest have been highlighted as:
● of special interest
●● of outstanding interest
1.Smyth, D.R., Bowman, J.L., Meyerowitz, E.M. Early flower development in Arabidopsis. Plant Cell 1990;2(8):755–767.
2.Mandel, M.A., Gustafson-Brown, C., Savidge, B., Yanofsky, M.F.. Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 1992;360(6401):273–277.
3.Shannon, S., Meeks-Wagner, D.R.. Genetic Interactions That Regulate Inflorescence Development in Arabidopsis. Plant Cell 1993;5(6):639–655.
4.Liljegren, S.J., Gustafson-Brown, C., Pinyopich, A., Ditta, G.S., Yanofsky, M.F.. Interactions among APETALA1, LEAFY, and TERMINAL FLOWER1 specify meristem fate. Plant Cell 1999;11(6):1007– 1018.
5.Kwiatkowska, D.. Flower primordium formation at the Arabidopsis shoot apex: quantitative analysis of surface geometry and growth. J Exp Bot 2006;57(3):571–580.
6.Kwiatkowska, D.. Flowering and apical meristem growth dynamics. J Exp Bot 2008;59(2):187–201.
7.Coen, E.S., Meyerowitz, E.M.. The war of the whorls: genetic interactions controlling flower development. Nature 1991;353(6339):31–37.
8.Alvarez-Buylla, E.R., Ben ́ıtez,
M., Corvera-Poiré, A., Chaos Cador, A., de Folter, S., Gamboa de Buen, A., et al.
Flower development. The Arabidopsis
book 2010;8:e0127. ●● In this review, the author summarizes the different published data about
Arabidopsis flower development. The author explains the
flower formation, the different genes that have a key role in this formation and
the interest of the genetic regulatory network models to understand the ABC
system.
9.Gustafson-Brown, C., Savidge, B., Yanofsky, M.F.. Regulation of the Arabidopsis floral homeotic gene APETALA1. Cell 1994;76(1):131–143.
10.Chen, X.. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science (New York, NY) 2004;303(5666):2022–5.
11.Zhao, L., Kim, Y., Dinh, T.T.,
Chen, X.. miR172 regulates stem cell fate and defines the inner
boundary of APETALA3 and PISTILLATA
expression domain in Arabidopsis floral meristems.
Plant J 2007;51(5):840–9. ● By using the effect of expression of miR172-resistant
AP2, Zhao shows the role of the MicroRNA miR172 in the
repression of AP2 in the two inner whorls of the flower.
miR172 acts in the same region than AG but largely independently of this gene
expression.
12.Honma, T., Goto, K.. The Arabidopsis floral homeotic gene PISTILLATA is regulated by discrete cis-elements responsive to induction and maintenance signals. Development (Cambridge, England) 2000;127(10):2021–30.
13.Goto, K., Meyerowitz, E.M.. Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Gene Dev 1994;8(13):1548–1560.
14.Sundström, J.F., Nakayama, N.,
Glimelius, K., Irish, V.F.. Direct regulation of the floral
homeotic APETALA1 gene by APETALA3 and
PISTILLATA in
Arabidopsis. Plant J
2006;46(4):593–600. ● Based on mutant observations of depletion or over-expression of different
genes, the author shows the regulation of AP1 by AP3 and PI proteins. AP3 and PI
play a key role in the down-regulation of AP1.
15.Espinosa-Soto, C.,
Padilla-Longoria, P., Alvarez-Buylla, E.R.. A gene regulatory
network model for cell-fate determination during Arabidopsis
thaliana flower development that is robust and recovers experimental
gene expression profiles. Plant Cell 2004;16(11):2923–2939. ●● The author summarizes published ABC and non-ABC gene interaction data in
logical tables to integrate in a dynamic network model. The model of 15 genes is
able to attain 10 attractors, which 4 explain the ABC stable pattern.
16.Drews, G.N., Bowman, J.L., Meyerowitz, E.M.. Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product. Cell 1991;65(6):991–1002.
17.Liu, Z., Meyerowitz, E.M.. LEUNIG regulates AGAMOUS expression in Arabidopsis flowers. Development 1995;121(4):975–91.
18.Deyholos, M.K., Sieburth, L.E.. Separable whorl-specific expression and negative regulation by enhancer elements within the AGAMOUS second intron. Plant Cell 2000;12(10):1799–810.
19.Gomez-Mena, C., de Folter, S.,
Costa, M.M.R., Angenent, G.C., Sablowski, R.. Transcriptional
program controlled by the floral homeotic gene AGAMOUS during
early organogenesis. Development 2005;132(3):429–38. ●● Based on mRNA expression, the author studies the AG interaction from the
onset of organogenesis. AG is expressed at the stage 3 in
the two inner whorls. It is involved in a positive-feedback loop with SEP3, AP3
and PI.
20.Melzer, R., Theissen, G.. Reconstitution of 'floral quartets' in vitro involving class B and class E floral homeotic proteins. Nucleic Acids Res 2009;37(8):2723–36.
21.Immink, R.G.H., Tonaco, I.a.N., de Folter, S., Shchennikova, A., van Dijk, A.D.J., Busscher-Lange, J., et al. SEPALLATA3: the 'glue' for MADS box transcription factor complex formation. Genome Biol 2009;10(2):R24.
22.Smaczniak, C., Immink, R.G.H.,
Blanvillain, R., Busscher, M., Busscher-lange, J., Liu, S., et al. Characterization of MADS-domain transcription factor complexes in
Arabidopsis flower development. P Natl
Acad Sci USA 2012; ●● AP1, AP3, PI, AG, SEP3 are MADS domain proteins. Based on
immunoprecipitation, the author shows the different interactions between these
proteins and with other proteins present in the flower. These interactions
explain the quartet model.
23.Urbanus, S.L., de Folter, S., Shchennikova, A.V., Kaufmann, K., Immink, R.G.H., Angenent, G.C.. In planta localisation patterns of MADS domain proteins during floral development in Arabidopsis thaliana. BMC Plant Biol 2009;9:5.
24.Krizek, B.A., Fletcher, J.C.. Molecular mechanisms of flower development: an armchair guide. Nat Rev Genet 2005;6(9):688–698.
25.Parcy, F., Nilsson, O., Busch, M.a., Lee, I., Weigel, D.. A genetic framework for floral patterning. Nature 1998;395(6702):561–6.
26.Conner, J., Liu, Z.. LEUNIG, a putative transcriptional corepressor that regulates AGAMOUS expression during flower development. Proc Natl Acad Sci USA 2000;97(23):12902–7.
27.Samach, A., Klenz, J.E., Kohalmi, S.E., Risseeuw, E., Haughn, G.W., Crosby, W.L.. The UNUSUAL FLORAL ORGANS gene of Arabidopsis thaliana is an F-box protein required for normal patterning and growth in the floral meristem. Plant J 1999;20(4):433–445.
28.Chae, E., Tan, Q.K.G., Hill,
T.a., Irish, V.F.. An Arabidopsis F-box
protein acts as a transcriptional co-factor to regulate floral
development. Development 2008;135(7):1235–45. ●● UFO, an F-box protein, is necessary to the activation of
AP3 expression. This protein, expressed at the stage 2
in the whorls 2 and 3, is shown to be a transcriptional co-factor with LFY that
regulates the expression of AP3.
29.Laufs, P.. Separable roles of UFO during floral development revealed by conditional restoration of gene function. Development 2003;130(4):785–796.
30.Gregis, V., Sessa, A.,
Dorca-Fornell, C., Kater, M.M.. The
Arabidopsis floral meristem identity genes
AP1, AGL24 and SVP directly repress class B and C floral
homeotic genes. Plant J 2009;60(4):626–37. ● few ABC and non-ABC genes play a key role at the very early stages of the
flower development (AP1, AGL24, SVP). The author shows that
class B and C genes are repressed by a co-repressor complex with LUG, SEU and
two dimers : AP1-AGL24 and AP1-SVP.
31.Heisler, M.G., Jönsson, H.. Modelling meristem development in plants. Curr Opin Plant Biol 2007;10(1):92–7.
32.Jönsson, H., Heisler, M., Reddy, G.V., Agrawal, V., Gor, V., Shapiro, B.E., et al. Modeling the organization of the WUSCHEL expression domain in the shoot apical meristem. Bioinformatics 2005;21 Suppl 1:i232–40.
33.Kauffman, S.. Homeostasis and Differentiation in Random Genetic Control Networks. Nature 1969;224(5215):177–178.
34.Aldana, M., Coppersmith, S., Kadanoff, L.. Boolean Dynamics with Random Couplings. Perspectives and Problems in Nonlinear Science 2002;93(4):23–90. 0204062.
35.Mendoza, L., Xenarios, I..
A method for the generation of standardized qualitative
dynamical systems of regulatory networks. Theor Biol Med Model 2006;3:13. ●● The author proposes a general method to study the genetic regulatory
network. Starting with a discrete method, he can easily find the different
steady states. Integrate these steady states in a continuous model of ordinary
differential equations allows to have good understanding of the
system.
36.Balleza, E., Alvarez-Buylla, E.R., Chaos, A., Kauffman, S., Shmulevich, I., Aldana, M.. Critical dynamics in genetic regulatory networks: examples from four kingdoms. PloS one 2008;3(6):e2456.
37.Hasty, J., Mcmillen, D., Isaacs, F., Collins, J.J.. Computational studies of gene regulatory networks. Genetics 2001;2(April).
38.Mendoza, L., Alvarez-Buylla, E.R.. Dynamics of the genetic regulatory network for Arabidopsis thaliana flower morphogenesis. J Theor Biol 1998;193(2):307–319.
39.Mendoza, L., Thieffry, D., Alvarez-Buylla, E.R.. Genetic control of flower morphogenesis in Arabidopsis thaliana: a logical analysis. Bioinformatics 1999;15(7-8):593–606.
40.Chaos, A. ,Aldana ,M. ,Espinosa-Soto, C., Léon, B.G.P., Arroyo, A.G .,Alvarez-Buylla, E.R.. From Genes to Flower Patterns and Evolution: Dynamic Models of Gene Regulatory Networks. J Plant Growth Reg 2006;25(4):278–289.
41.Alvarez-Buylla, E.R., Chaos, A.,
Aldana, M., Ben ́ıtez, M., Cortes-Poza, Y., Espinosa-Soto, C., et al. Floral morphogenesis: stochastic explorations of a gene network
epigenetic landscape. PloS one 2008;3(11):e3626. ● Starting with the boolean genetic regulatory network of 15 genes of the
Arabidopsis flower, the author shows that noise can
explain the transition between the different attractors. The proximity of these
attractors is the same in this model than in the flower ABC system.
42.Sanchez-Corrales, Y.E.,
Alvarez-Buylla, E.R., Mendoza, L.. The Arabidopsis
thaliana flower organ specification gene regulatory network
determines a robust differentiation process. J Theor Biol
2010;264(3):971–83. ●● The author proposes at first the last updated genetic regulatory network
of the Arabidopsis flower development based on the last
published data. Using a continuous model, the author finds 14 transitional
steady states that correspond to transitory stages in the apparition of the four
floral attractors.
43.Barrio, R.A., Hernandez-Machado,
A., Varea, C., Romero-Arias, J.R., Alvarez-Buylla, E.. Flower
development as an interplay between dynamical physical fields and genetic
networks. PloS one 2010;5(10):e13523. ● Based on the last genetic regulatory network of the
Arabidopsis flower, the author gives an explanation of
the space ABC pattern establishment by modelisation by partial differential
equations of few gene interactions and the existence of a ”phase-field” coupled
with a ”spontaneous curvature-like” term.
44.Ming, Y.. A model for floral organ development in Arabidopsis. 2009;:1–13.
45.Benıtez, M., Espinosa-Soto, C., Padilla-Longoria, P., Alvarez-Buylla, E.R.. Interlinked nonlinear subnetworks underlie the formation of robust cellular patterns in Arabidopsis epidermis: a dynamic spatial model.BMC Syst Biol 2008;2:98.
46.Chisholm, R.H., Hughes, B.D., Landman, K.a.. Building a morphogen gradient without diffusion in a growing tissue. PloS one 2010;5(9).
47.Irish, V.F.. The flowering of Arabidopsis flower development. Plant J 2010;61(6):1014–28.
48.Giorgianni, M.W., Mann, R.S.. Establishment of medial fates along the proximodistal axis of the Drosophila leg through direct activation of dachshund by Distalless. Dev Cell 2011;20(4):455–68.


