Evolution of gene network controlling plant reproductive development
2012/11/01
Abstract
Understanding the phylogeny of the homeotic genes that control floral organ identities, their regulators and interactions, may help us to understand the key steps in the evolution of reproductive mechanisms in the land plants, leading ultimately to the flower. The regulatory network that controls flower development comprises genes that regulate floral organ identity, most of which belongs to the MADS-box family, and their master regulator, LEAFY. In this review, Idescribe the evolution of plant reproductive structures from freshwater charophycean algae through bryophytes, ferns, gymnosperms and angiosperms, and our current knowledge of the MADS-box and LEAFY-like genes that control reproductive development in the land plants and their closest algal relatives.
Table of Contents
- Introduction
- The angiosperm flower: its morphology and gene regulatory network
- Evolution of plant reproduction: from green algae to flowers
- LEAFY evolution
- Gene duplications in the MADS-box family correlate with growing complexity in reproductive anatomy in the land plants
- Conclusion
- References and recommended reading
Introduction
Plants are a vital part of our planetary ecosystem without which there would be neither oxygen nor food to support animal life. When most people think of plants, they think of flowering plants. However, flowering plants are only the latest in a long line of major plant life forms. The land plants also include bryophytes (liverworts, mosses and hornworths), ferns and gymnosperms (conifers, etc). These groups show a progressive increase in physical size and complexity in their reproductive systems, leading finally to the angiosperms.
Within the angiosperms, which today comprise some 400 000 species, three basally diverging lineages can be recognized. These lineages, collectively termed the "ANA-grade" (for Amborellales, Nymphaelales, Austrobaileyales), appear to have conserved many likely ancestral features of the first flowers, and hence may prove useful to studies of the origin of the flowering plants.
Much is already known of the molecular control of flower development in model flowering plants such as Arabidopsis thaliana (Arabidopsis) and comparative studies are beginning to explain how a network of developmental control genes has been modified to generate progressively more elaborate and efficient reproductive structures throughout land plant evolution.
The purpose of this report is to retrace the evolutionary history of reproductive development in the plant kingdom, and the gene regulatory network that controls it, from the algal ancestors of land plants to model flowering plants.
The angiosperm flower: its morphology and gene regulatory network
The shoot, or above-ground part of an angiosperm, typically consists of a system of radially symmetrical stems, which grow indeterminately from groups of meristematic cells at their apices. These apical meristems also produce lateral structures, typically including leaves, for photosynthesis and flowers, which function in sexual reproduction [1].
Despite the enormous variation which exists in flower size, shape, symmetry and pigmentation, the basic organization of the flower is remarkably conserved between species. Four organ types are typically present, arranged in concentric whorls (Figure 1). From the outside to the inside of the flower, these organs are typically: sepals, which protect the flower bud; petals, which may function to attract insect pollinators; stamens, which produce the pollen; and finally carpels, which contain the ovules and later form the tissues of the fruit [1].
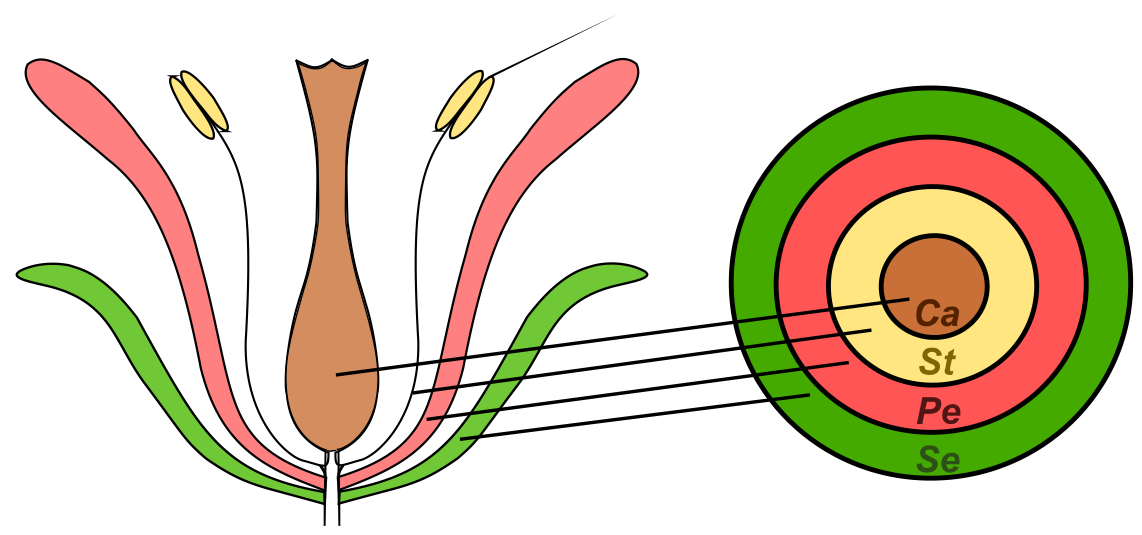
Figure 1. Schematic representation of a typical angiosperm flower containing organs arranged in concentrated whorls. .
Ca – carpels, St – stamens, Pe – petals, Se – sepals.
Each floral organ type is determined by a unique combination of four gene activities, called A, B, C and E. Accordingly, A+E activity specifies sepals, A+B+E activity specifies petals, B+C+E activity specifies stamens and C+E activity specifies carpels [2]. In Arabidopsis, all except one of the genes of this "ABCE Model" encode type II MIKC MADS-box transcription factors (the one exception being APETALA2, an A-class protein that encodes a distinct AP2- class transcription factor). Other MADS-box floral homeotic genes of the ABCE model include: the A-class gene APETALA1 (AP1), two B-class genes APETALA3 (AP3) and PISTILLATA (PI), the C-class gene AGAMOUS (AG) and four E-class genes, SEPALLATA1-4 [2]. A D-class of floral homeotic gene function has also been defined. D-class genes are closely related to C-class genes and share C-activity in some species, while also contributing to ovule identity [3,4].
MADS-box genes are found in a wide range of eukaryotes including metazoans, fungi, slime molds, and green plants, though type II MIKC MADS-box factors have diverged in plants [5]. These factors contain MADS-box (M), intervening (I), keratin-like (K), and C-terminal (C) domains (reviewed in [2,5]). The MADS-box domain is responsible for binding to "CArG" motifs in DNA, while the I- and K-domains function in homo- and/or heterodimer formation. Floral homeotic MADS-box proteins are believed to bind to pairs "CArG" motifs. Such binding involves the formation of tetramers of MADS-box proteins, which takes place through interactions involving their C-terminal domains [2]. Accordingly, in Arabidopsis, the four types of floral organs are specified by tetramers of different compositions of A, B, C and E proteins, yielding the "Floral quartet model" [2].
In Arabidopsis, flowering is initiated by a combination of environmental and endogenous signals, which result in the transition of the shoot apical meristem which produces leaves at its flanks, to an inflorescence meristem, which produces floral meristems at its flanks [6]. LEAFY (LFY) is the key regulator of patterning in the floral meristem. This gene encodes a transcription factor that acts as a master controller by activating the genes of the "ABCE model" in the appropriate regions of the floral meristem [6‑9], as shown in Figure 2. Moreover, AP1 can activate LFY, PI induces AP3 expression and vice versa (reviewed in [10]) and A- and C-class proteins negatively regulate each other, which explains why no floral whorl contains both A- and C-activity (for reviews of "ABCE model" see [5,10]). To generate correct floral patterning, LFY requires the cofactors UNUSUAL FLORAL ORGANS (UFO), which is necessary for AP3 induction in the second and third whorls [11] and WUSCHEL (WUS), which results in AG expression in the third and fourth whorls [10]. SEP genes are also direct targets of LFY, and the proteins they encode are present in the MADS-box tetramers that specify all floral organ types [2,12].
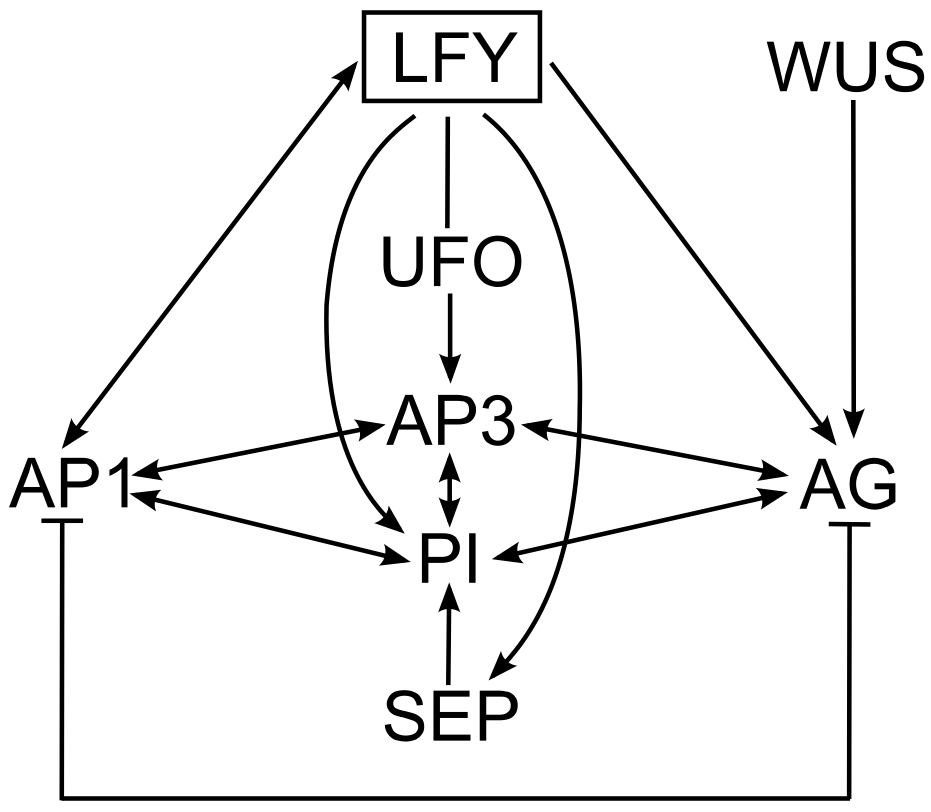
Figure 2. The gene regulatory network controlling flower development in Arabidopsis. .
The arrows represent direct inductions. Main regulator: LFY – LEAFY, cofactors: UFO – UNUSUAL FLORAL ORGANS and WUS – WUSCHEL, A-class gene: AP1 – APETALA1, B-class genes: AP3 – APETALA3 and PI – PISTILLATA, C-class gene: AG – AGAMOUS, E-class gene: SEP – SEPALLATA.
Evolution of plant reproduction: from green algae to flowers
Plant life began in water and to colonize the terrestrial environment, plants had to adapt to much dryer conditions. These adaptations affected not only the morphology and physiology of their vegetative organs, but also the processes and organs of reproduction.
The closest relatives of the land plants are charophycean freshwater green algae [13]. In these algae, male gametes produced in antheridia are free to migrate through the aquatic milieu to fertilize female gametes produced in oogonia [13,14]. The life cycle of charophyceans is haplobiontic, in which the zygote represents the only diploid cell during the life cycle [1].
Even if bryophytes were the first plants to colonize the land and their diploid sporophyte produces a single type of dessication-resistant haploid spore, their reproduction is still dependant on the water. Haploid gametes are produced in antheridia and archegonia on the gametophyte and fuse only in amoist environment to form adiploid zygote which undergoes mitosis. All land plants are characterized by an alternation of two life forms: a haploid gametophyte generation that produces gametes, and adiploid sporophyte generation growing from an embryo, that produces spores. The spore, first present in bryophytes, was a key novelty for reproduction in the relatively dry terrestrial environment, because it is easily dispersed, allowing the bryophytes to colonize the land surface [1].
Water dependance of the gametophyte persists in lycophytes and ferns which evolved after the bryophytes. The sporophyte generation became dominant and continues to develop from a zygote produced in the tissues of the gametophyte. The sporophyte produces spores as a means of dissemination and resistance to dessication. Haploid spores are produced in the sporangia on the abaxial (under) side of reproductive leaves. Moreover, two extant genera of lycophytes and a few leptosporangiate ferns are characterized by heterospory. These plants produce two types of spores: male microspores and female megaspores, in respective micro- and megasporangia, which develop into microgametophytes with antheridia and megagametophytes with archegonia. Heterospory persists also in further emerging lineages of land plants, because it promotes outcrossing which results in the long- term evolutionary advantage of better gene-flow within populations [1].
Vascular systems, firstly evolved in lycophytes, promoted an extensive growth of the sporophyte generation and its independance of the gametophyte in subsequent lineages of land plants which grow in relatively dry environment. In contrast, the tiny haploid gametophyte became dependant on the dominant diploid sporophyte. Seeds, which appeared in the first seed plants, contains an embryo sporophyte, enclosed in tissues of the previous sporophyte generation to protect it from dessication. Two main clades of plants can be distinguished in seed plants: the gymnosperms which have naked ovules and seeds, and the angiosperms, in which the ovules are formed within a closed structure, termed the carpel.
In gymnosperms, the female and male gametophytes develop individualy inside the ovules and microsporangia, respectively. The female gametophyte situated in the megasporangium or nucellus is surrounded by one protective integument, or seed-coat. These three parts make up the ovule. Each ovule is typically located on an ovuliferous scale and subtended by a bract, creating a megasporophyll. The megasporophylls are typically circularly attached on the seed cone axis. The male gametophyte is contained within each pollen grain, and is composed of two or three nuclei: one vegetative and one or two generative developing to sperm cells. After pollination and fertilization, a seed develops on megasporophylls of the seed cone until its dissemination [1]. Gnetales, a group of extant gymnosperms possess morphologically similar reproductive structures as angiosperms, but they evolved in parallel with the flower of flowering plants [15].
The angiosperm flower appears to have evolved from the seed and pollen cone structures. In general, angiosperm flowers contain both male and female organs. Thus, the gene regulatory network controlling flower development had to change significantly from the gymnosperm network controlling the development of separate unisexual structures.
Angiosperms are the most recent group of extant land plants to appear. The "ABCE model" of floral organ identity was based mainly on studies of Arabidopsis and Antirhinnum majus, which have a relatively advanced and conserved floral structure. In contrast, angiosperms from the early-diverging "ANA-grade" typically display a simpler and less uniform flower structure and various different taxa exhibite: an undifferentiated perianth composed of tepals, a well-differentiated perianth composed of sepals and petals, or no perianth [1]. By investigating the gene network that controls reproductive development in all groups of land plants and their closest aquatic relatives, we can identify the molecular changes that finally led to the evolution of flower.
LEAFY evolution
LEAFY (LFY) protein is a regulator integrating the environmental and autonomous signals that lead to flowering [16]. Until now, no LFY homologue has been found in freshwater charophycean algae, similar to the ancestors of the land plants. In the moss Physcomitrella patens, two LFY-like genes have been identified. The LFY-like gene products are important for the first cell division of the zygote and may regulate the expression of genes responsible for zygote development [17]. However, none of the targets of LFY in Arabidopsis respond to LFY-like proteins from P. patens in transgenic Arabidopsis plants [18]. Therefore, two hypothetic scenarios can be considered for the functional evolution of LFY among the land plants. In the first scenario, LFY might control similar or slightly different gene network in non-flowering and flowering plants. In this case, co-evolution of cis-acting motifs in target genes and of DNA-binding specificity in LFY would result in the inability of bryophyte LFY to regulate angiosperm targets, and vice versa. Alternatively, there might have been a complete change of LFY function between non-flowering and flowering plants in which a change in biochemical activity recruited new LFY targets [18].
The expression pattern of two LFY homologues, which were identified in the fern model Ceratopteris richardii, supports the hypothesis of progressive evolution of a gene network controlling reproductive development, with LFY-like protein as a main regulator. The LFY-like genes are expressed in tissues including shoot tips and circinate reproductive leaves [19]. However, no Arabidopsis floral homeotic gene can be induced by fern LFY-like proteins in transgenic Arabidopsis plants. Thus, we have to consider the possibility that LFY targets in C. richardii might be considerably different from those of Arabidopsis [18].
The LFY-like gene duplicated before the gymnosperm and angiosperm divergence (Figure 3). In gymnosperms, LFY-like proteins induces the floral homeotic gene homologs [18], but the function of each duplicate slightly differs. The first identified gymnosperm LFY-like gene was NEEDLY (NLY) from Pinus radiata. After the transition to the reproductive phase, NLY‘s expression pattern and regulatory network is very similar to that of angiosperm LFY. However, early NLY expression in reproductive structures occurs mostly in female structures [20]. The second LFY-like gene in P. radiata is expressed similarly in both female and male cones. Thus, the presence of two LFY-like paralogues in conifers might separate reproductive determination of individual male and female reproductive organs [21]. Angiosperm LFY is a transcription factor with a proline-rich domain at its amino terminus and an acidic region in the middle part of the protein. Surprisingly, gymnosperm LFY-like protein lacks these domains. The absence of these domains might be a cause of evolutionary changes that took place after the divergence of the angiosperm and gymnosperm lineages [20].
Gene duplications in the MADS-box family correlate with growing complexity in reproductive anatomy in the land plants
Only one MADS-box gene containing M-, I-, K-and C-domains has been found and characterized in each of the investigated charophycean green algae. In these algae, MADS-box gene expression occurs during gametangium differentiation and decreases after fertilization. Thus, MADS-box genes are associated with reprodutive development even in the closest external relatives of the land plants [22]. The presence of a keratin-like (K) domain in algal MADS-box proteins, which is responsible for the dimerization in Arabidopsis, suggests that MADS-box proteins had been recruited to form dimers or quaternary complexes before the origin of the land plants (Figure 3).
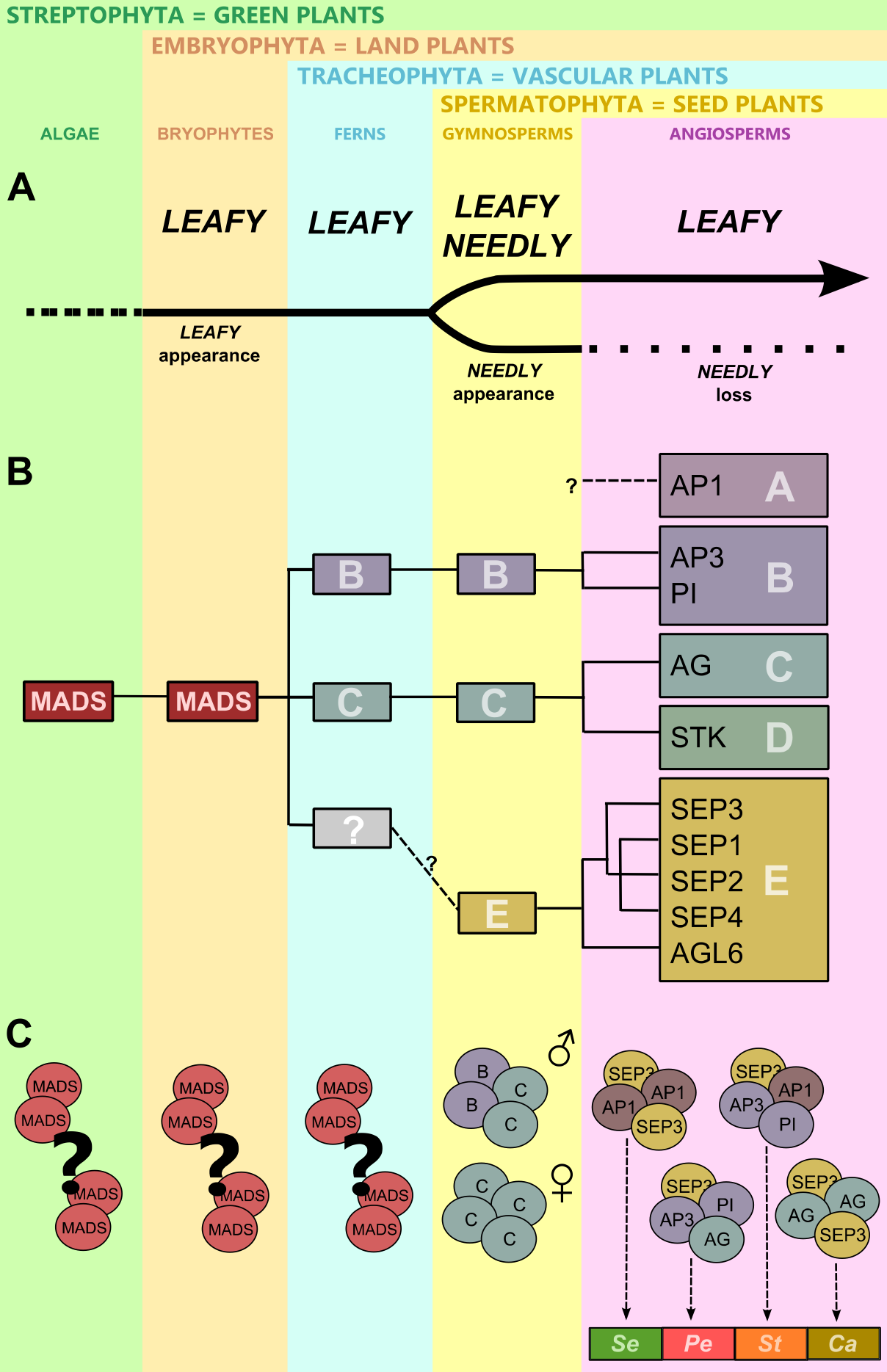
Figure 3. Evolution of the gene regulatory network controlling plant reproduction. .
(A) Appearance, a major duplication and a loss of LEAFY-like genes. (B) Major duplications of MADS-box family genes. A – A-class genes: eg AP1 – APETALA1; B – B-class genes: eg AP3 – APETALA3 and PI – PISTILLATA; C – C-class genes: eg AG – AGAMOUS; D – D-class genes: eg STK – SEEDSTICK; E – E-class genes: SEP1-4 – SEPALLATA1-4 and AGL6 – AGAMOUS LIKE 6. (C) Possible dimerization and tetramer formation in algae, bryophytes and ferns. Formation of quaternary complexes specifying male and female reproductive organs in gymnosperms and reproductive organ identities in angiosperms, Se – sepals, Pe – petals, St – stamens, Ca – carpels.
In contrast to the situation in charophycean algae, MADS-box genes are abundantly present in all land plants, including those in flowering plants that specify floral organ identities. The common ancestor of vascular plants and mosses possessed at least two types of MADS-box gene, so the type II MIKC MADS-box gene duplication must have occured before the separation of mosses and vascular plants, more than 450 MYA [23].
MADS-box genes described in the moss model P. patens [23] possess a putative promoter region. This region seems to be controlled not only by M-domain proteins, because they contain putative "CArG" motifs [24], known to bind M-domain proteins, but also by LFY-like proteins because of its consensus LFY-like protein binding site (CCANTG) [9]. Thus, the MADS-box genes in mosses might be involved in complex gene regulatory networks similar to those in flowering plants [23].
From the fern model C. richardii five MADS-box genes from three subfamilies were cloned. They are expressed in the meristematic regions and primordia of sporophyte shoots and roots, as well as in reproductive leaves and sporangial initials. The lack of organ-specific expression of MADS-box genes in fern reproductive structures, resembling that of floral homeotic genes in flowering plants, may indicate that the restriction of MADS-box gene expression to specific reproductive organs took place later in the evolution of MADS box genes [25]. The undifferentiated expression of fern MADS-box genes might also be related to the fact that ferns have simpler reproductive organs than seed plants. Thus, the evolution of MADS-box genes expression may result from the evolution of genes regulating MADS-box genes (eg. LFY) [19]. Moreover, Arabidopsis MADS-box genes from the B- and C-classes have some conserved motifs of amino acid residues which had been found also in fern MADS-box genes (reviewed in [26]). Thus, we can suggest that ferns have orthologs of angiosperm B- and C-class genes which might be associated with the evolution of heterospory that is the forerunner of male and female reproductive organs in seed plants.
The direct induction of MADS-box genes by LFY-like protein well-known in seed plants has been established to have originated after the divergence of ferns from seed plants lineage and before the divergence of gymnosperms and angiosperms [19]. Since LFY-like genes induce floral homeotic gene homologs in seed plants, the induction of MADS-box genes by a LFY-like gene might be a key event in the evolution of floral homeotic genes from generally expressed MADS-box genes [19]. An ancient "BC system" was already established in the most recent common ancestor of extant gymnosperms and angiosperms about 300MYA [27]. Gymnosperm B- and C-class genes have similar functions to their angiosperms orthologs. The expression of B- and C-class genes together specifies the identity of pollen- bearing organs [28] and the C-class gene expression alone specifies seed-cone development [29].
However, any ortholog of the SEP and A-class genes required for floral organ and meristem identity determination has not been detected in gymnosperms [30,31]. In contrast, AGL6-like genes, which are similar to the SEP1-4-like genes, has been characterized in gymnosperms. Gymnosperm AGL6-like expression is similar to that of angiosperm SEP1-4 genes in reproductive organs: it overlaps the expression of B- a C-class genes [32,34]. Thus the common ancestor of AGL6-like and SEP-like genes might have had a function in floral-like organ determination [33] and in the synchronisation of B- and C-class gene expression for the determination of male and female organ identity [34].
In Gnetophyta, agroup of gymnosperms whose reproductive structures superficially ressemble those of the flowering plants, the mechanism for the determination of reproductive organ identity operates similarly to that in flowering plants. In vitro, Gnetophyta orthologs of B-class and C-class proteins together, or C-class proteins alone, can loop DNA in tetramers similar to the angiosperm floral quartet complexes. In vitro, these Gnetum MADS-box proteins do not require other factors such as E-class floral homeotic proteins to „glue“ them together in multimeric complexes, suggesting that the evolutionary origin of floral quartet formation might not depend on E-class proteins [35]. However, the situation might be different in vivo.
In contrast to the gymnosperm clade, basal "ANA-grade" angiosperms possess homologs of all MADS-box gene A-, B-, C-, D- and E-classes. The majority of theses genes are expressed in the floral organs of "ANA-grade" angiosperms similarly to their Arabidopsis orthologues. For example, in basal angiosperms AP3 and PI homologs are expressed in tepals and stamens, while AG homologs are expressed in stamens and carpels. SEP3 homologs in basal angiosperms are expressed in all floral organs similarly to SEP3 in Arabidopsis, suggesting this gene to perform similar functions throughout the angiosperm clade. However, some floral identity genes have additional expression domains in "ANA-grade". For example, expression of the AG and PI homologs mentioned above was detected also in the carpels, and that of AG homologs in the perianth, of some basal angiosperm taxa. Furthermore, homologs of the A-class gene AP1 show strong expression throughout all floral organs in "ANA-grade" angiosperms. Thus, the expression pattern of these genes has a conserved component as well as a broader component, compared to the "ABCE model" [36].
Conclusion
MADS-box genes are associated with the development of reproductive structures within all clades of extant land plants and their freshwater algal relatives. Their extensive duplication and subsequent specification led to the growing complexity of the reproductive structures. MADS-box genes duplicated several times and each duplicate specified its function in different parts of the reproductive meristem to assure the development of different reproductive organs. LFY, as the master regulator of floral homeotic MADS-box genes in angiosperm regulatory network, did not duplicate very much. LFY gene is present only in one or two copies. Thus, LFY may evolved by progressive changes in its DNA sequence together with changes in its biochemical activity. The presence of putative LFY-binding domain in the promoter region of some bryophyte MADS-box genes and the hypothesis of the possible LFY/MADS-box gene co-evolution might lead to a discovery of new LFY targets from ferns or bryophytes that would might be orthologs of the angiosperms ABCE genes. Moreover, by investigating the ancestors of proteins from "ABCE model" we could clarify the origin of higher order complexes from "Floral quartet model" and prove or disprove that gymnosperm E-class protein AGL6 functions similarly as Arabidopsis SEP to "glue" the floral homeotic gene products homologs together.
References and recommended reading
Papers of particular interest have been highlighted as:
● of special interest
●● of outstanding interest
1. Bold HC, Alexopoulos CJ, Delevoryas T: Morphology of Plants and Fungi.Harper International Edition; 1980.
2. Theissen G: Development of floral organ identity: stories from the MADS house.Current opinion in plant biology 2001, 4:75-85.
3. Favaro R, Pinyopich A, Battaglia R, Kooiker M, Borghi L, Ditta G, Yanofsky MF, Kater MM, Colombo L: MADS-Box Protein Complexes Control Carpel and Ovule Development in Arabidopsis. The Plant cell 2003, 15:2603-2611.
4. Pinyopich A,
Ditta GS, Savidge B, Liljegren SJ, Baumann E, Wisman E, Yanofsky MF:
Assessing the redundancy of
MADS-box genes during carpel and ovule development.Nature 2003,
424:85-8. ● Author showed the existence of AG-independent carpel-development
pathway which converts sepals to carpeloid organs even in the
ag mutant, leading to the establishment of D-class
genes specifying carpels and ovules development.
5. Theissen G, Becker a, Di Rosa a, Kanno a, Kim JT, Münster T, Winter KU, Saedler H: A short history of MADS-box genes in plants.Plant molecular biology 2000, 42:115- 49.
6. Sablowski R: Flowering and determinacy in Arabidopsis.Journal of experimental botany 2007, 58:899-907.
7. Parcy F, Nilsson O, Busch MA, Lee I, Weigel D: A genetic framework for floral patterning. October 1998, 395:561-566.
8. Wagner D, Sablowski RW, Meyerowitz EM: Transcriptional Activation of APETALA1 by LEAFY.Science 1999, 285:582-584.
9. Busch MA, Bomblies K, Weigel D: Activation of a Floral Homeotic Gene in Arabidopsis. Science 1999, 285:585-587.
10. Alvarez-Buylla ER, Azpeitia E, Barrio R, Benítez M, Padilla-Longoria P: From ABC genes to regulatory networks, epigenetic landscapes and flower morphogenesis: Making biological sense of theoretical approaches.Seminars in cell & developmental biology 2010, 21:108-17.
11. Chae E, Tan QK-G, Hill T a, Irish VF: An Arabidopsis F-box protein acts as a transcriptional co-factor to regulate floral development.Development 2008, 135:1235-45.
12. Ditta G, Pinyopich A, Robles P, Pelaz S, Yanofsky MF: The SEP4 Gene of Arabidopsis thaliana Functions in Floral Organ and Meristem Identity.Current Biology 2004, 14:1935-1940.
13. Lewis LA, McCourt RM: Green algae and the origin of land plants.American Journal of Botany 2004, 91:1535-56.
14. Niklas KJ, Kutschera U: The evolution of the land plant life cycle.The New phytologist 2010, 185:27-41.
15. Shindo S, Ito M, Ueda K, Kato M, Hasebe M: Characterization of MADS genes in the gymnosperm Gnetum parvifolium and its implication on the evolution of reproductive organs in seed plants.Evolution & development 1999, 1:180-90.
16. Parcy F: Flowering: a time for integration.The International journal of developmental biology 2005, 49:585-93.
17. Tanahashi T, Sumikawa N, Kato M,
Hasebe M: Diversification of gene function: homologs of the floral
regulator FLO/LFY control the first zygotic cell division in the moss
Physcomitrella patens.Development 2005, 132:1727-36. ● These study describes the function of FLO/LFY homologs in the moss model
Physcomitrella patens which significantly differs from
the LFY-like genes function in angiosperms.
18. Maizel A,
Busch M a, Tanahashi T, Perkovic J, Kato M, Hasebe M, Weigel D:
The floral regulator LEAFY evolves
by substitutions in the DNA binding domain.Science 2005,
308:260-3. ●● This paper showed that Arabidopsis LEAFY protein
do induce no MADS-box genes controlling reproductive development in
mosses or ferns and very small number in gymnosperms. Moreover,
LEAFY-like homologs from mosses or ferns are
less
active in Arabidopsis transgenic
assays suggesting the progressive evolution of LFY and its targets or
complete change of LFY function between basal taxa and flowering
plants.
19. Himi S, Sano
R, Nishiyama T, Tanahashi T, Kato M, Ueda K, Hasebe M: Evolution of MADS-box gene induction by FLO/LFY
genes.Journal of molecular evolution 2001, 53:387-93. ● The authors demonstrated the duplication and subsequent loss of FLO/LFY
homologs during vascular plants evolution and the expression pattern of
the Ceratopteris
richardii LFY-like gene
suggesting that MADS-box genes are not induced by LFY-like protein at
the stage of ferns.
20. Mouradov A, Glassick T, Hamdorf B, Murphy L, Fowler B, Marla S, Teasdale RD: NEEDLY, a Pinus radiata ortholog of FLORICAULA/LEAFY genes, expressed in both reproductive and vegetative meristems.Proceedings of the National Academy of Sciences of the United States of America 1998, 95:6537-42.
21. Mellerowicz EJ, Horgan K, Walden A, Coker A, Walter C: PRFLL - a Pinus radiata homologue of FLORICAULA and LEAFY is expressed in buds containing vegetative shoot and undifferentiated male cone primordia.Planta 1998, 206:619-29.
22. Tanabe Y,
Hasebe M, Sekimoto H, Nishiyama T, Kitani M, Henschel K, Münster T, Theissen G,
Nozaki H, Ito M: Characterization
of MADS-box genes in charophycean green algae and its implication for the
evolution of MADS-box genes.Proceedings of the National
Academy of Sciences of the United States of America 2005, 102:2436-2441. ● These study describes MADS-box genes and their
expression pattern in charophycean algae suggesting the implication of
MADS-box genes in the reproductive development already in the closest
relatives of land plants.
23. Henschel K, Kofuji R, Hasebe M,
Saedler H, Münster T, Theissen G: Two ancient classes of MIKC-type
MADS-box genes are present in the moss Physcomitrella patens.
Molecular biology and evolution 2002, 19:801-14. ● The authors characterized seven MADS-box genes from the moss model
Physcomitrella patens and showed that the common
ancestor of bryophytes and other vascular plants had at least two copies of
MADS-box gene which evolved to MIKCc- and MIKC*-type of MADS-box
genes.
24. Schwarz-Sommer Z, Hue I, Huijser P, Flor PJ, Hansen R, Tetens F, Lönnig WE, Saedler H, Sommer H: Characterization of the Antirrhinum floral homeotic MADS-box gene deficiens: evidence for DNA binding and autoregulation of its persistent expression throughout flower development.EMBO J. 1992, 11:251-63.
25. Hasebe M, Wen CK, Kato M, Banks JA: Characterization of MADS homeotic genes in the fern Ceratopteris richardii.Proc. Natl. Acad. Sci. U. S. A. 1998, 95:6222-7.
26. Hasebe M: Evolution of Reproductive Organs in Land Plants.Journal of Plant Research 1999, 112:463-474.
27. Winter KU, Becker a, Münster T, Kim JT, Saedler H, Theissen G: MADS-box genes reveal that gnetophytes are more closely related to conifers than to flowering plants.Proceedings of the National Academy of Sciences of the United States of America 1999, 96:7342-7.
28. Sundström J, Engström P: Conifer reproductive development involves B-type MADS-box genes with distinct and different activities in male organ primordia.The Plant Journal 2002, 31:161-9.
29. Zhang P, Tan HTW, Pwee K-H, Kumar PP: Conservation of class C function of floral organ development during 300 million years of evolution from gymnosperms to angiosperms.The Plant Journal 2004, 37:566-577.
30. Becker A: The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Molecular Phylogenetics and Evolution 2003, 29:464-489.
31. Theissen G, Melzer R: Molecular mechanisms underlying origin and diversification of the angiosperm flower. Annals of botany 2007, 100:603-19.
32. Mouradov A, Glassick T, Hamdorf B, Murphy L, Marla S, Yang Y, Teasdale R: Family of MADS-Box genes expressed early in male and female reproductive structures of monterey pine.Plant physiology 1998, 117:55-61.
33. Melzer R, Wang Y-Q, Theissen G: The naked and the dead: the ABCs of gymnosperm reproduction and the origin of the angiosperm flower.Seminars in cell & developmental biology 2010, 21:118-28.
34. Carlsbecker A, Tandre K, Johanson U, Englund M, Engström P: The MADS-box gene DAL1 is a potential mediator of the juvenile-to-adult transition in Norway spruce (Picea abies).The Plant Journal 2004, 40:546-57.
35. Wang Y-Q, Melzer R, Theissen
G: Molecular interactions of orthologues of floral homeotic
proteins from the gymnosperm Gnetum gnemon provide a clue to
the evolutionary origin of “floral quartets”.The Plant Journal 2010, 64:177-190. ●● This study demonstrated in vitro formation of B-
and C-class protein ternary complexes specifying male and female reproductive
structures in gymnosperm species Gnetum gnemon suggesting
the possible evolution of angiosperm floral quartets.
36. Kim S, Koh J, Yoo M-J, Kong H, Hu Y, Ma H, Soltis PS, Soltis DE: Expression of floral MADS-box genes in basal angiosperms: implications for the evolution of floral regulators.The Plant Journal 2005, 43:724-44.


