Biological Mechanisms of Noise in Gene Expression
2012/10/01
Abstract
How could we explain that genetically identical cells such as clones are not totally identical? They differ by the rate of their gene expression. This phenomenon is particularly due to non- deterministic fluctuations, or noise, of biological mechanisms. Noise can be divided into two classes. Extrinsic noise arises from fluctuations of the environment, from inevitable variations in the random partitioning of molecules between daughter cells when cells divide but also from the heterogeneity of cell size and shape and cell cycle stage. Intrinsic noise is due to inherent stochasticity of biochemical processes such as transcription and translation. Thus, it has been demonstrated that proteins are produced in dynamic rate because mRNAs are transcribed in pulses or bursts. Noise propagation can be affected by regulatory circuits which can either be detrimental for cells or confer a selective advantage on them.
Table of Contents
Introduction
Any individual in a population of living organisms or cells is unique. Most inter-individual phenotypic variabilities are due to genetic differences. So how could we explain that clones are not totally identical? Environment and history seem to contribute in variability in cellular phenotypes. Over the last few years, many scientists have tried to explain that phenomenon. Now, we know that clones for instance differ by the rate of their gene expression, as defined by the set of reactions controlling the abundance of gene products. The term of noise or stochasticity in gene expression is commonly used to refer to the measured level of variation in gene expression among isogenic cells (i.e., genetically identical), grown homogeneously in a common environment. To date, four potential sources of noise have been described [1]:
- “The random nature of chemical reactions within a cell;
- The differences in the internal states of a population of cells, either from a predictable process such as cell cycle progression or from random processes such as partitioning of mitochondria during cell division;
- Subtle environment differences;
- Genetic mutations”.
Here, we propose to review the molecular sources of the inherent stochasticity of gene expression and to describe some characteristics of noise. We will try to define the particular relation that can exist between noise and regulatory networks and how, in some cases, that can lead to the emergence of diseases. This review will probably be helpful to discuss the role of noise in the evolution.
Extrinsic noise versus intrinsic noise
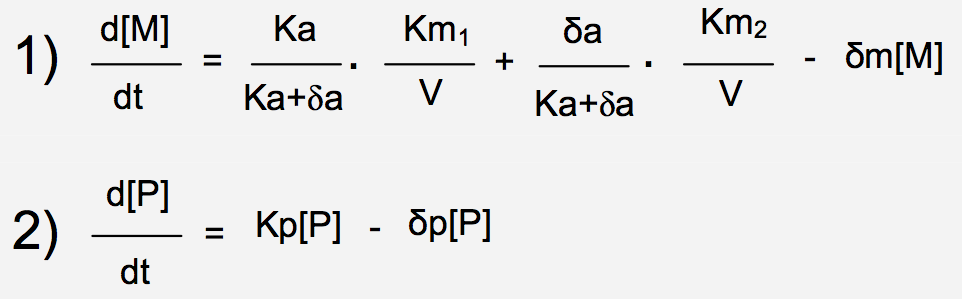
Stochasticity in gene expression has been suggested some years ago to be the source of cell-to- cell variations among isogenic populations [2]. Thus far, it was difficult to determine experimentally whether the variation in the product of a given gene came from fluctuations in cellular components that could lead indirectly to variation in expression of the gene [3] or noise in expression of the gene itself. Elowitz et al., used the model of Escherichia coli to better understand the sources of noise. They constructed strains that enable the discrimination of the two types of noise: extrinsic and intrinsic (Figure 1) [4].
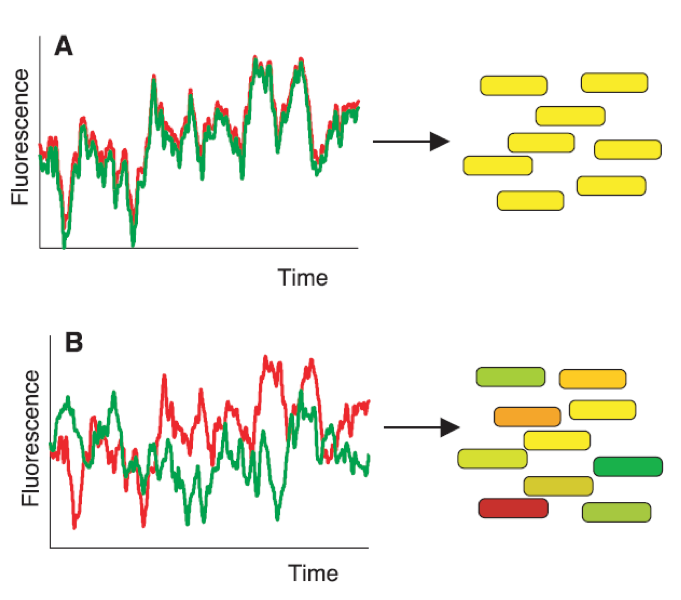
Figure 1. Two sources lead to variations in gene expression: intrinsic and extrinsic noise..
Elowitz et al. [4] constructed strains by integrating two reporter genes, cfp (green) and yfp (red) controlled by identical promoters. Those bacteria enable the discrimination of two sources of noise: extrinsic noise leads to cells expressing the same amount of both protein and appear in yellow (A) whereas intrinsic noise leads to cells expressing different amount of each proteins and appear in red or green. (Reproduced from Elowitz et al. [4]).
Then, extrinsic noise has been better characterized. It arises from fluctuations of the environment, from inevitable variations in the random partitioning of molecules between daughter cells when cells divide but also from the heterogeneity of cell size and shape and cell cycle stage. Those elements are global for a single cell but vary from one cell to another. To explain the relationship between the noise and the size of a cell, Kœrn et al,. took the example of a protein that can freely move from the cytoplasm to the nucleus [5]. At equilibrium, both the cytoplasm and the nucleus concentrations are equal but if one single protein translocates into the nucleus, then the consequences onto the nucleus concentration would be stronger than consequences of a protein translocating into the cytoplasm because the volume of the latter is bigger. Additionally, it has been shown that extrinsic noise can also arise from variations in the amount of transcriptional activators common to all genes such as RNA polymerase II [6]. Regarding cell cycle, extrinsic fluctuations can be reduced by isogenic cell synchronization.
Now, if we consider a totally theoretical population of cells that are identical not only genetically but also in the concentration and state of their cellular components, then such population would still vary from cell-to-cell because of the random and inherent stochasticity of biochemical processes such as transcription and translation [4]. Because most of the reagents that participate in these chemical reactions are present in extremely low concentration in cells, noise in chemical reactions is inherent. Raser and O'Shea measured the intrinsic noise strength of various yeast promoters at different rate of expression and demonstrate that noise in gene expression does not always depend on the rate of expression [7]. Indeed, GAL1 and PHO84 promoters exhibit a low level of intrinsic noise which does not vary with a variation on the rate of expression. Inversely, PHO5 promoter exhibits a larger intrinsic noise which decreases while increasing the rate of gene expression. Moreover, intrinsic noise seems to be promoter-specific: even if PHO84 and PHO5 have opposite noise characteristics, they share a common transcriptional regulator.
Thus, intrinsic noise is an important source of random fluctuation because of its key role in variations among isogenic populations.
Intrinsic noise: a particular framework
Over the last years, research on intrinsic stochasticity has converged to a unique framework to explain gene expression noise (Figure 2 and Box 1). That framework can be divided into three steps. First, proteins are produced in dynamic rate rather than in a uniform rate because mRNAs are transcribed in bursts. Secondly, at protein level, the burst is buffered due to their long lifetime. Finally, noise produced by a given gene can propagate.
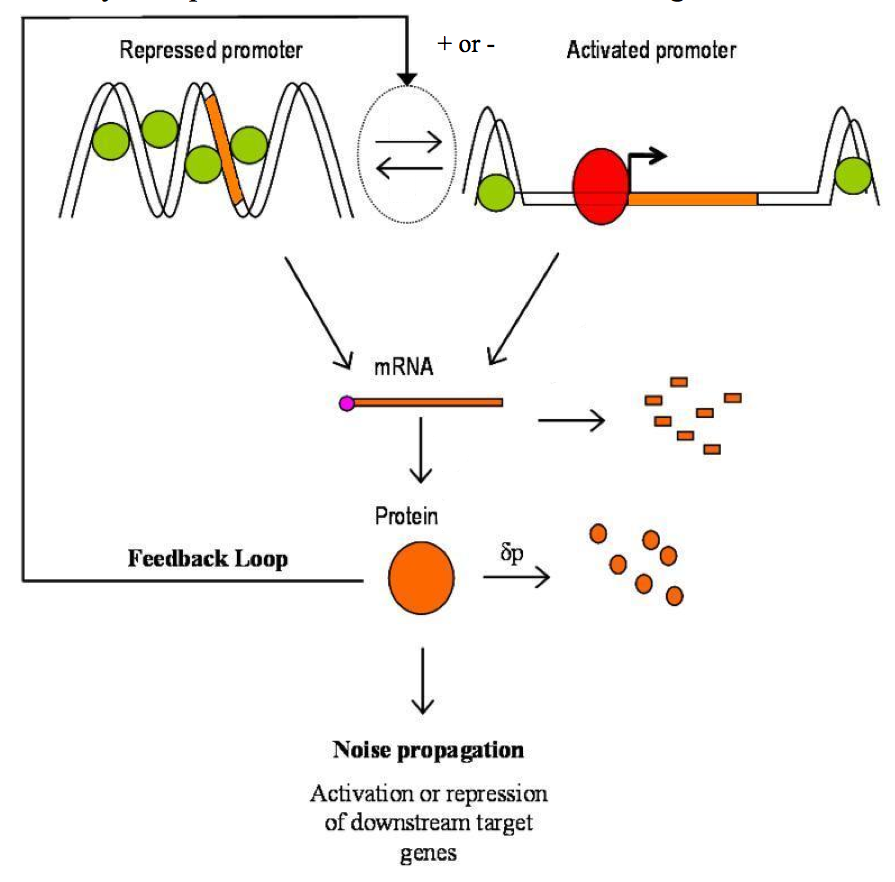
Figure 2. Intrinsic noise: a particular framework..
Intrinsic noise can be divided into three steps: first mRNAs are produced in bursts because of stochastic transitions from repressed promoter state to activated promoter state and vice versa. Secondly, the burst production of proteins is buffered because of their slow degradation rate. Finally, noise propagates through because the level of proteins expression can influence downstream target genes. Feedback loops can exist. Positive ones tend to increase the noise whereas negative feedback loops tend to reduce it. (Adapted from Kœrn et al. [5]).
Transcriptional noise
It has been further demonstrated that transcription of individual genes in eukaryotic cells occurred stochastically and infrequently. This means that for any given gene, mRNAs are produced in pulse or burst, reflecting the inherently stochastic nature of gene expression [8]. Three kinetics mechanisms of promoter transcriptional activation has been described (Figure 3) [7]:
- Case 1) “The activation step is infrequent compared to the transcription and the active promoter is stable”.
- Case 2) Identical to the Case 1 but the active promoter is unstable.
- Case 3) “The activation step is frequent compared to transcription and the activated promoter is highly unstable”.
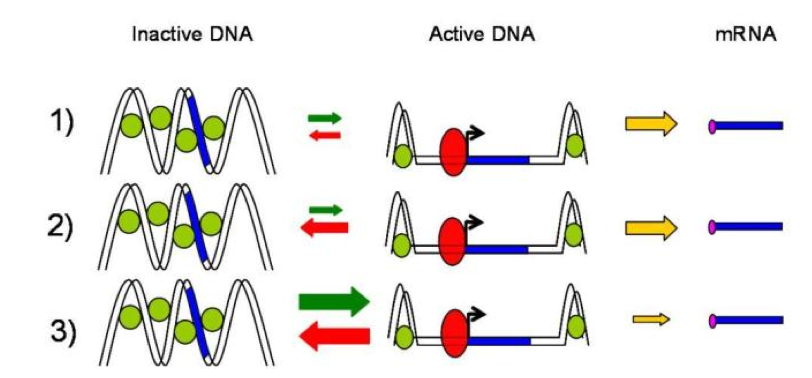
Figure 3. Three kinetics mechanisms of promoter transcriptional activation. .
Case 1) “The activation step is infrequent compared to the transcription and the active promoter is stable”. That could correspond to a promoter that is activated or inactivated by slow chromatin remodeling in which nucleosomes are remove or replace on the DNA. Case 2) Identical to the Case 1 but the active promoter is unstable. That could correspond to a faster and more reversible activation step associated with nucleosome sliding. Case 3) “The activation step is frequent compared to transcription and the activated promoter is highly unstable. That event could be associated with rapid activator binding-dissociation reactions in which transcription occurs for a fraction of the binding events”. That third case seems to be more likely seen in prokaryotes. (Adapted from Raser JM and O'Shea EK [1]).
Raser et O'Shea performed stochastic simulations by varying whether promoter activation step or transcriptional efficiency in order to see how a change in the steady-state mean of gene expression could affect the intrinsic noise strength. They showed that in case 1, promoter activation strongly decreases the noise strength whereas this latter strongly increases while increasing transcriptional efficiency. For case 2, the profile is roughly the same but varying these kinetic constants affects the noise strength much less. The case 3 seems to be the one which produces the less noise regardless of variations.
These stochastic simulations underlie three means for a cell to up-regulate its transcription: by increasing the rate of gene activation; increasing the rate of transcription when the gene is in an active state and finally, by decreasing the rate of gene inactivation [6].
Since experimental techniques allow the distinction between extrinsic and intrinsic noise, many studies have been done to determine whether stochastic gene expression originating from mRNAs bursts could be controlled by biological mechanisms.
The study of the PHO5 yeast promoter enabled to classify that promoter into the case 1 noise strength profile [7]. It has been hypothesized that at the inactive state, PHO5 promoter displays positioned nucleosomes. The binding of the Pho4 transcription factor to upstream activating sequences UAS1 and UAS2 allow the recruitment of chromatin- remodeling complexes that remove nucleosomes away from PHO5 promoter which becomes accessible for transcription. Indeed, mutations in these UAS sites prevent the nucleosomal disruption leading to a less efficient PHO5 activation. At the same time, the two PHO5 UAS mutants have a higher strength noise compared to the wild type, which promoter activation decreases the noise strength. Remodeling PHO5 promoter requires multiple chromatin remodeling complexes such as SWI/SNF, INO80 and SAGA [9-11].
To test further the hypothesis that PHO5 promoter activation step requires chromatin remodeling, the noise strength after induction of PHO5 promoter was measured in yeast mutants lacking one of these three chromatin remodeling complexes. All resulted in increased noise strength consistent with hypothesis made in case 1 stochastic model
Ansel et al. used the inheritable characteristic of noise to further investigate its genetic control [12]. Indeed, they supposed that if noise was controlled by genetic mechanisms, then it should be heritable through yeast strains generations. They used two yeast strains with different background showing different levels of noise from Met17 promoter whereas the mean expression of that gene was identical. They introduce GFP gene under the expression of Met17 promoter at the HIS genomic locus in segregants issued from a cross between these two strains and put in evidence that both noise and mean were under genetic control. After screening, they identified a QTL (Quantitative trait loci) from position 116330 to 207819 on chromosome V that has been found to confer high noise level in yeast. In that locus, the presence of URA3 gene was tested to determine whether that gene has a role in noise modulation. It has been shown that noise in ura3∆0 mutant lacking the entire URA3 gene was larger than in wild type yeast. Thus, URA3 seems to play an important role in decreasing noise. It is important to notice that these genetic mechanisms controlling noise level involve trans-regulation because none of them were located near the HIS3 or Met17 genes.
Ura3∆0 mutation leads to an inhibition of uracil synthesis; the pool of nucleotides available for RNA synthesis is then reduced [13]. Thus, the authors investigated whether transcriptional elongation was involved into noise. To this aim, they measured the noise level of dst1/ppr2∆0 yeast mutant. DST1/PPR2 normally encodes the transcription elongation factor SII (TFIIS) that stimulates transcript elongation by binding to RNA polymerase II and facilitating its passage through intrinsic pausing sites in vitro [14]. They observed a dramatic increase of noise meaning that impairing transcriptional elongation can increase the gene expression noise. Thus, their study revealed that a QTL including URA3 and DST1 genes can control the level of noise by decreasing it.
Other sites such as TATA box sequences, which are dispensable for chromatin remodeling but important for transcriptional efficiency, seem to play a role in increasing the noise strength. This phenomenon has been particularly observed for the PHO5 yeast promoter activation [7].
The TATA box sequences play an important role in assembling the transcription machinery at promoters [15].Transcription activation is coordinated with the binding of TATA binding protein (TBP) onto TATA box consensus sequences through interactions with coactivators complexes such as TFIID (Transcription factorIID) or SAGA (Spt-Ada-Gcn5-Acetyltransferase [16-18]. Then, these complexes recruit the RNA polymerase II that starts the transcription. In yeast, only 20% of total genes contain a TATA box. However, TATA-less promoters also require TBP for function [19]. By searching for new TATA-like consensus sequences, Basehoar et al., found that there are two types of genes in yeast: the TATA box-containing genes that are highly regulated and associated with response to stress and the TATA-less promoters, more likely associated with housekeeping genes [20]. The former preferentially uses SAGA rather than TFIID used by the latter.
A study made on the TATA containing SAGA regulated PDR5 gene, that encodes a protein related to the large ABC family of transporters [21], has demonstrated that genes regulated by the coactivator SAGA are likely to be transcribed in pulse. Indeed, it seems that at least in yeast, many genes showing high variation in protein level are regulated by SAGA and contain a highly conserved TATA box sequences [22,23]. Finally, even in mammalian cells, gene expression is subject to large and intrinsic fluctuations. To investigate the mechanisms controlling transcriptional bursts in CHO cells (Chinese hamster ovary), Raj et al., altered the global level of transcription both by changing the amount of transcriptional activators present in cells and by changing the number of binding sites for that activator [6]. In this aim, the YFP gene was inserted downstream a minimal cytomegalovirus promoter. Upstream of that promoter were inserted one or seven copies of tetracycline tet operator sequence allowing the transcription only when a tet transactivator protein binds to the operator sequence. The tet transactivator protein can be prevented to bind to DNA by using tetracyclin-like antibiotic doxycycline. Thus, it becomes possible to control the level of free tet transactivator in cells by varying the concentration of doxycycline (Figure 4).
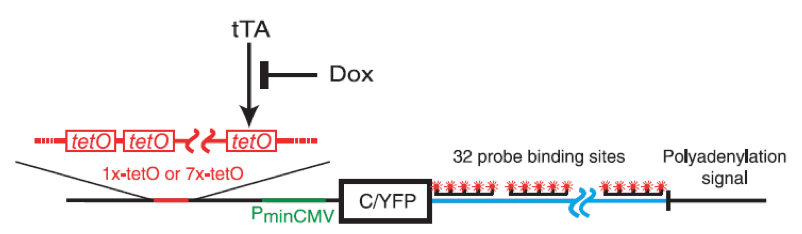
Figure 4. In mammalian cells, gene expression is subject to large and intrinsic fluctuations..
Schematic diagram of the YFP gene controlled by one or seven copies tetracycline tet operator sequences. The transcription is possible only when tet transactivator protein tTA binds to the operator sequence. Doxycycline binds to tTA preventing the transcription. (From Raj et al. [6]).
They found that increasing either transcriptional factor binding sites or the amount of transcriptional activators in cells increased the average burst size rather than their frequency. Nevertheless, it was impossible to determine relative to the framework explaining gene expression noise, whether that increase in burst size was due to a decrease of gene inactivation state or an increase of gene activation state.
The long lifetime of proteins acts as a buffer
A theoretical model based on yeast suggests that “frequent transcription followed by inefficient translation results in lower intrinsic noise in protein levels than does infrequent transcription followed by efficient translation” (Figure 5) [1]. Moreover, it has been proposed that yeasts can adopt two strategies to produce a given amount of any proteins [24,25]: (1) They can maximize their transcription and minimize their translation per mRNA. This leads to low stochasticity due to a minimization of noise at protein level. (2) A maximization of translation per mRNA correlated with a minimization of transcription will result in larger noise due to high translation.
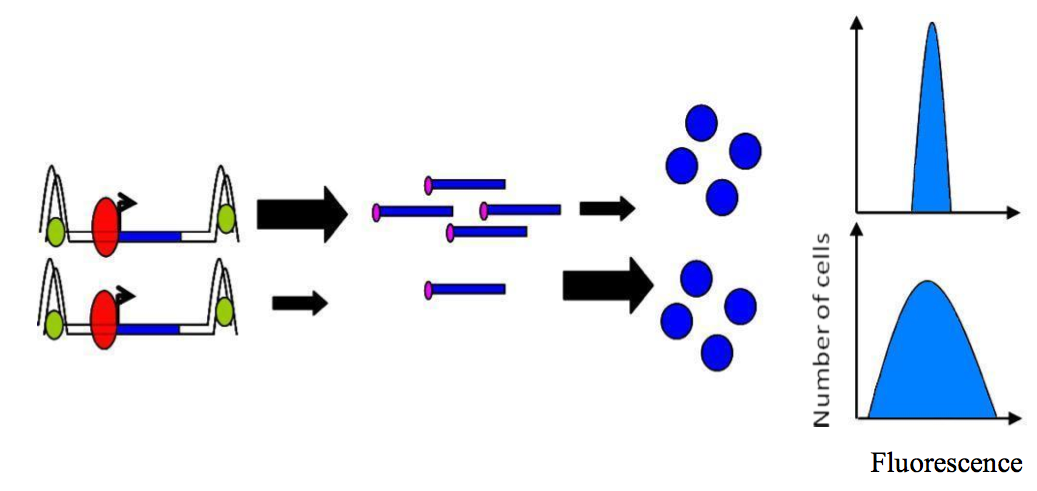
Figure 5. Control of noise..
Frequent transcription followed by inefficient translation (top) results is lower intrinsic noise than infrequent transcription followed by efficient translation (bottom). (Adapted from Raser JM and O'Shea EK [1]).
So, we could imagine for instance in the case of some essential proteins, that fluctuations in their level could be more detrimental to cells than fluctuations in the level of other proteins. Essential genes (i.e., genes which depletion onto both alleles is lethal) would have high transcription rates associated with low number of translation per mRNA so that noise could be minimized. It is important to notice that there is a correlation between gene's dispensability (as defined as “the growth defect of a yeast strain missing that gene in rich glucose medium” [24]) and its rate of protein production. This means that essential genes are more tightly controlled than non essential ones. In order to test whether essential genes tend to adopt preferentially the first strategy regarding to non essential genes, Fraser et al., classified more than 4,000 genes into 15 bins dependently of their protein production rate. Bins were then separated into three classes depending on their number of translation per mRNA. It appeared that most of the essential genes have significantly both the higher transcription rate and the lower translation rate leading in low noise at protein level. Those results were confirmed mathematically [23] and in the same time, studies have shown that noise level is even lower for haploinsufficient genes (i.e., dose-sensitive genes, defined as “genes that reduce growth when their rate is decreased by half in heterozygotes”) [26,27].
Essential proteins and dosage-sensitive genes are not the only ones exhibiting low noise level. Fraser et al., also took an interest on proteins that participate in stable protein complexes. Genes encoding them also have high level of transcription per mRNA and low level of translation. Genes encoding subunits of protein complexes have to be tightly regulated; producing too little or too more of a subunit could compromise the assembly of the complex. Thereby, controlling noise for essential genes or genes encoding complex subunits can prevent a waste of energy for cells.
Nevertheless, it has been shown in mammalian cells that burst in gene expression could be buffered at protein level by slow protein degradation rate [6]; this happens when the proteins halftime is longer than that of mRNAs. That phenomenon makes difficult the analysis of subtle intrinsic noise which often requires the use of long halftime fluorescent proteins.
Noise propagation
Living cells use complex networks composed of interacting genes and proteins to implement various cellular and developmental programs. These network architectures are difficult to study because they depend on cellular states and on cell context. Gene regulations occur with a delay. Indeed, one must take into account that protein concentrations have to be sufficient to have a regulatory effect on downstream target genes. Such delay does not occur for extrinsic noise because it affects all genes over time. So, by following the expression of multiple genes over time in individual cells, it becomes possible to discriminate extrinsic noise correlations from regulatory correlations (correlation could be defined as protein concentrations of the multiple genes observed). Thus, in order to further understand noise correlations, several studies have been done by introducing simple synthetic gene circuits into the Escherichia coli model [28,29].
Dunlop et al. used stochastic modeling and differentiated extrinsic noise which is global to all measured genes from intrinsic noise that leads to genes fluctuating independently from each others [29]. Their construct was done by using the bacteriophage λ CI repressor fused to yellow fluorescent protein (YFP). CI represses the production of red fluorescent protein (RFP) which is fused to the λ Pr promoter. On the same plasmid, a gene encoding cyan fluorescent protein (CFP) was fused to a constitutive promoter that is independent of CI repressor as a control of extrinsic noise. Thereby, CFP was expressed at homogeneous level among cells whereas a strong anti-correlation was observed between CI-YFP and RFP meaning that the RFP concentration is inversely proportional from YFP concentration over time. Mathematical modeling revealed that with only extrinsic noise, these three signals were positively correlated although YFP represses RFP. On the other hand, with only intrinsic noise, the repression of RFP by YFP leads to an anti-correlation between the two genes. Thus, in regard of such synthetic system introduced into E.coli, intrinsic noise is preponderant to extrinsic noise.
These observations highlight that the rate of expression of a given gene is largely influenced by the level of expression of upstream transcription factors that are themselves subject to transcriptional bursting. Thus, noise propagates from one gene to downstream target genes.
Feedback loop
The topology of regulatory circuits can sometimes reduce noise propagation. It was shown in some bacteria that noise associated with positive feedback could create phenotypic heterogeneity [30-32]. It has been shown among an isogenic population of Mycobacterium tuberculosis that such association could create a fraction of cells that are resistant to some stresses such as oxygen or nutrient deprivation making the pathogens able to survive in a latent state. In mycobacteria, the expression of rel, a protein involved in stress response, initiates a stringent response leading to persistence. Sukera et al. observed that rel expression is bistable meaning that there are two stable expression states for rel gene, low and high [32]. Gene expression noise plays an important role in driving the transition from low to high expression state. Thus, the combination of positive feedback and noise can have positive effects on the evolution of a population.
In Bacillus subtilis, Maamar et al. took an interest on the auto-stimulatory positive feedback loop in which ComK proteins, encoded by the comK gene, promote their own production [30]. That protein is involved in the competence of B. subtilis allowing them to uptake foreign DNA that increases their fitness. Uptaking foreign DNA in stress conditions increases the probability to get a resistant gene. ComK expression is also bistable; in one state the expression of ComK protein is low and the positive autoregulatory loop is not activated, in the other state, ComK concentration exceeds a critical threshold that activates the positive loop. That transition occurs during the stationary phase of growth. The authors demonstrated that intrinsic noise arising from that gene was responsible to the transition to competence due to large fluctuations at stationary phase that activate the positive feedback loop. Decreasing intrinsic noise in comK expression leads to a dramatic decrease of competent cells.
Conversely, negative feedback loops have been showed to decrease transcriptional noise [33,34].
Conclusion
Along this paper, we review some characteristics of noise. We put in evidence the existence of two types of genes; those encoding proteins forming multicomponent complexes, or dosage-sensitive genes and essential ones that tend to have low gene expression noise, and stress-related genes responding to changes in the environment that exhibit high noise level. In the latter case, the variability in protein content among cells can confer a selective advantage.
Nevertheless, noise can be detrimental to organism fitness. For instance, in diseases linked to haploinsufficience, increasing intrinsic noise could result in a total loss of function as presumed in the case of the tumor suppressor gene NF1 [35]. Another example is the onset of autosomal dominant diseases that could emerge later than on birth by the increasing of noise at the protein level [36]. The study of noise in gene expression could also help us to better understand why, in cancer, such particular mutations not always lead to the development of the disease. Thereby, Ansel et al., proposed to revisit the interpretation of incomplete penetrance because in cases of pathologies triggered by single-cells, mutations could lead to an increase in stochastic fluctuations allowing the emergence of some deviant phenotypic cells [12].
Finally, low noise level could have been selected for some genes in order to prevent harmful stochastic variations that could be deleterious for cells. In return, that phenomenon limits the ability of these genes to respond to perturbations. Nevertheless, overall stochastic fluctuations are probably one of the main ways, along with genetic mutations, that evolution has found to derive beneficial population diversity.
References and recommended reading
Papers of particular interest have been highlighted as:
● of special interest
●● of outstanding interest
1.Raser JM and O'Shea EK. Noise in gene expression: origins, consequences and control.
Science 2005, 309:2010-2013. ●● This review summarizes noise terminology and comment on recent
investigations into the sources, consequences, and control of noise in gene
expression.
2.McAdams HH and Arkin A. It's a noisy business! Genetic regulation at the nanomolar scale. Trends Genet 1999, 5:65-9.
3.Swain PS, Elowitz MB and Siggia ED. Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc Natl Acad Sci USA 2002, 99:12795-12800.
4.Elowitz MB, Levine AJ, Siggia ED and Swain PS. Stochastic gene expression in a single cell. Science 2002, 297:1183-1186. ● In this paper, authors constructed strains of Escherichia coli that enable detection of noise and discrimination between extrinsic and intrinsic noise.
5.Kœrn M, Elston TC, Blake WJ and Collins JJ. Stochasticity in gene expression: from theories to phenotypes. Nature 2005, 452:451- 464.
6.Raj A, Peskin CS, Tranchina D, Vargas DY and Tyagi S. Stochastic mRNA synthesis in mammalian cells. Plos Biol 2006, 4:1707-1719. ● The authors found that there are massive variations in the number of mRNA molecules present in each cell because mRNAs are synthesized in bursts.
7.Raser JM and O'Shea EK. Control of stochasticity in eukaryotic gene expression. Science 2004, 304:1811-1815. ● In this paper, the authors identified mutations that alter the noise of gene expression. These mutations suggest that noise is an evolvable trait.
8.Blake WJ, Kœrn M, Cantor CR and Collins JJ. Noise in eukaryotic gene expression. Nature 2003, 422:633-637. ● Authors used Saccharomyces cerevisiae to show that noise arises from transcription and contributes to the level of heterogeneity within a eukaryotic clonal population. They also explore the propagation of noise in a gene cascade network and demonstrate experimentally that increased noise in the transcription of a regulatory protein leads to increased cell-cell variability in the target gene output.
9.Steger DJ, Haswell ES, Miller AL, Wente SR and O'Shea EK. Regulation of chromatin remodeling by inositol polyphosphates. Science 2003, 299:114-120.
10.Barbaric S, Reinke H and Hörz W. Multiple mechanistically distinct functions of SAGA at the PHO5 promoter. Mol Cell Biol 2003, 23:3468-3476.
11.Barbaric S, Walker J, Schmid A, Svejstrup JQ and Hörz W. Increasing the rate of chromatin remodeling and gene activation--a novel role for the histone acetyltransferase Gcn5. EMBO J 2001, 20:4944-4951.
12.Ansel J, Bottin H, Rodriguez-Beltran C, Damon C, Nagarajan M, Fehrmann S, François J and Yvert G. Cell-to-cell stochastic variation in gene expression is a complex genetic trait. PLoS Genet 2008, 4:e1000049. ● This paper demonstrates the existence of a QTL including URA3 and DST1 genes that can control the level of noise in yeasts.
13.Mason PB and Struhl K. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol Cell 2005, 17:831-840.
14.Kipling D and Kearsey SE. Function of the S. cerevisiae DST1/PPR2 gene in transcription elongation. Cell 1993, 72:12.
15.Benoist C and Chambon P. In vivo sequence requirements of the SV40 early promotor region. Nature 1981, 290:304-310.
16.Agalioti T, Chen G and Thanos D. Deciphering the transcriptional histone acetylation code for a human gene. Cell 2002, 111:381-392.
17.Hassan AH, Prochasson P, Neely KE, Galasinski SC, Chandy M, Carrozza MJ and Workman JL. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 2002, 111:369- 379.
18.Lemon B and Tjian R.Orchestratedresponse: a symphony of transcription factors for gene control. Genes Dev 2000, 14:2551-2569.
19.Pugh BF and Tjian R. Transcription from a TATA-less promoter requires a multisubunit TFIID complex. Genes Dev 1991, 5:1935-1945.
20.Basehoar AD, Zanton SJ and Pugh BF. Identification and distinct regulation of yeast TATA box-containing genes. Cell 2004, 116:699-709. ● The authors found that there are two types of genes in yeast: the TATA box-containing genes that are highly regulated and associated with response to stress and the TATA-less promoters, more likely associated with housekeeping genes.
21.Leonard RC, Rodger A and Dixon JM.ABC of breast diseases. Metastatic breast cancer. BMJ 1994, 309:1501-1505.
22.Zenklusen D, Larson DR and Singer RH. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat Struct Mol Bio 2008, 15:1263-1271. [19011635] ● This paper demonstrated that at least in yeast, many genes showing high variation in protein level are regulated by SAGA and contain a highly conserved TATA box sequences.
23.Newman JR, Ghaemmaghami S, Ihmels
J, Breslow DK, Noble M, DeRisi JL and Weissman JS. Single-cell
proteomic analysis of S. cerevisiae reveals the architecture of
biological noise. Nature 2006, 441:840-846. ● This paper shows that noise in protein expression is dominated by the
stochastic production/destruction of mRNAs. Moreover, there are protein-specific
differences in noise that are strongly correlated with a protein's mode of
transcription and its function.
24.Fraser HB, Hirsh AE, Giaever G, Kumm J and Eisen MB. Noise minimization in eukaryotic gene expression. PLOS Biology 2004, 2:834- 838. ● In this paper, the authors estimated the noise in protein production for nearly every yeast gene, and demonstrated that the production of essential and complex-forming proteins involves lower levels of noise than does the production of most other genes.
25.Thattai M and van Oudenaarden A. Intrinsic noise in gene regulatory networks. Proc Natl Acad Sci USA 2001, 98:8614-9.
26.Lehner B. Selection to minimise noise in living systems and its implications for the evolution of gene expression . Mol Syst Biol 2008, 4:170. ● This paper shows that most of the essential genes have significantly both the higher transcription rate and the lower translation rate leading in low noise at protein level. And that noise level is even lower for haplo-insufficient genes.
27.Batada NN and Hurst LD. Evolution of chromosome organization driven by selection for reduced gene expression noise. Nat Genet 2007, 39:945-954. ●● This paper demonstrates that genes are organized in clusters. Essential gene clusters are associated with low nucleosome occupancy whereas non-essential genes clusters exhibit higher noise level.
28.Pedraza JM and van Oudenaarden
A. Noise propagation in gene networks.
Science 2005, 307:1965-1974. ●● To quantify how noise propagates through gene networks, the authors
measured expression correlations between genes in single cells. They found that
intrinsic noise was transmitted from upstream genes to downstream target
genes.
29.Dunlop MJ, Cox RS 3rd, Levine
JH, Murray RM and Elowitz MB. Regulatory activity revealed by
dynamic correlations in gene expression noise. Nat Genet
2008, 40:1493-1501. ●● In this paper, the authors use the bacteriophage λ that can affect
E.coli to demonstrate that intrinsic noise is
preponderant to extrinsic noise. This paper also helps to better understand
noise propagation.
30.Maamar H, Raj A and Dubnau D.
Noise in gene expression determines cell fate in
Bacillus subtilis. Science
2007, 317:526-535. ●● In Bacillus subtilis the authors took an interest
on the auto-stimulatory positive feedback loop involving ComK proteins. They
show that activation of this feedback loop can confer a selective advantage in
stress conditions.
31.Süel GM, Kulkarni RP, Dworki nJ, Garcia-Ojalvo J and Elowitz MB. Tunability and noise dependence in differentiation dynamics. Science 2007, 315:1716-1725.
32.Sureka K, Ghosh B, Dasgupta A, Basu
J, Kundu M and Bose I. Positive feedback and noise activate the
stringent response regulator rel in mycobacteria. PLOS One
2008, 3:e1771. ● This paper shows how, in Mycobacterium tuberculosis,
the combination of positive feedback and noise can have positive effects on the
evolution of a population.
33.Dublanche Y, Michalodimitrakis K, Kümmerer N, Foglierini M and Serrano L. Noise in transcription negative feedback loops: simulation and experimental analysis. Mol Syst Biol 2006; 2:41. [16883354]
34.Becskei A and Serrano L. Engineering stability in gene networks by autoregulation. Nature 2000, 405:590-593.
35.Kemkemer R, Schrank S, Vogel W, Gruler H and Kaufmann D. Increased noise as an effect of haploinsufficiency of the tumor-suppressor gene neurofibromatosis type 1 in vitro. Proc Natl Acad Sci USA 2002, 99:13783-13791.
36.Bosl WJ and Li R. The role of noise and positive feedback in the onset of autosomal
dominant diseases. BMC Syst Biol 2010, 4:93. ●● This paper shows that an increase of noise level can be responsible of
the onset of autosomal dominant diseases that could emerge later than on
birth.