Narcolepsy is a sleep disorder associated with polymorphisms in genes involved in immune response
21 - 01 - 2014
Abstract
Narcolepsy is a sleep disorder characterized by excessive daytime sleepiness, cataplexy and other associated symptoms. The prevalence of this disease is around 0.02% but is heterogenous between populations. Narcolepsy onset is during childhood or early puberty. This sleep disorder is caused by a loss of 70 000-90 000 hypocretin neurons representing at least 90% of the hypocretin cells pool. Narcolepsy is an immune system-associated disease linked with numbers of genes expressed in immune cells. More than 90% of narcoleptic patients are positive for HLA-DQB1*06:02 , an allele of the HLA class II. Narcolepsy is also associated with other genes involved in the immune response such as T-cell receptor α and Cathepsin H . All these genetic polymorphisms linked with narcolepsy may result in a strong immune response leading to an autoimmune process and the destruction of hypocretin neurons.
Table of Contents
- Introduction
- Narcolepsy is associated with polymorphism of genes implicated in antigen presentation by APC
- TCR is genetically linked with narcolepsy
- Narcolepsy is associated with genes involved in the survival and the proliferation of T-cells and the immunosuppressive effect of regulatory T-cells.
- Conclusion
- Acknowledgments
- References and recommended reading
Introduction
Narcolepsy is a sleep disorder characterized by five symptoms [1,2]: excessive daytime sleepiness, cataplexy (episodic loss of muscle ton), sleep paralysis, hypnagogic hallucinations (dream-like sensation near the onset of sleep) and disturbed nocturnal sleep. Many of the narcolepsy symptoms are explained by abnormalities of a particular stage of sleep, rapid eye movement (REM) sleep [2]. Narcolepsy prevalence is around 0.02% but is heterogenous between populations. In Japanese population, the prevalence reaches 0.16% whereas it is almost absent in Israeli population [3]•. Narcolepsy usually appears during childhood or early puberty [2]. When the five symptoms are present, the diagnosis is easy. Nevertheless if cataplexy or sleep paralysis are absent, there are two tests to confirm narcolepsy: the multiple sleep latency test (MSLT) and the polysomnogram [1][3]•. The MSLT is a sleep disorder diagnostic tool which measures the time taken by a patient to go to sleep in a quiet environment [4]. The test reveals that narcoleptic patients fall asleep more rapidly than controls (usually less than 8 minutes) [2]. The polysomnogram is an overnight test which takes continuous multiple measurements of many body functions as the brain or the muscle functions: it reveals abnormalities in REM sleep. Narcoleptic patients present a clear sleep stage dissociation with an increase number of REM and awake stages [2,5].
Narcolepsy is caused by an hypocretin deficiency. The first evidence linking hypocretin (also called orexine) with narcolepsy came from mutant animal models with defective hypocretin or hypocretin receptor 2 genes [6]••. In humans, narcolepsy is characterized by a loss of 70000-90000 hypocretin neurons of the hypothalamus [1][3]•. This loss represents 90% of the hypocretin neurons pool. Hypocretin neurons synthesize hypocretin which is a neurotransmitter involved in the regulation of wakefulness, appetite, and arousal [7]. Loss of hypocretin neurons seems to be sufficient to trigger narcolepsy [8]. Recently, the measure of hypocretin levels in cerebrospinal fluid have been considered as a diagnosis tool [9,10]. This test is more accurate than the MSLT and the polysomnogram as it directly addresses the biochemical cause of the disorder [9].
Narcolepsy is associated with upper airways infections such as influenza A virus and Streptococcus pyogenes infections [11]••[12]•. Indeed, a Chinese study conducted in 629 patients suggests a strong relationship between narcolepsy development and seasonal influenza infections [11]••. This study shows that narcolepsy incidence increases between April and August and is significantly correlated with seasonal H1N1 infections occurring during winter. It also reports that 2009 H1N1 pandemic has led to a 3-fold increase of narcolepsy incidence. Regarding Streptococcus pyogenes , specific markers of the infection (antibodies against β hemolytic streptococcus and Dnase B) are statistically higher in narcoleptic patients than in controls and their levels are even higher in case with recent onset [12]•. Nevertheless, the association between the sleep disorder and Streptococcus pyogenes infections is still controversial as no other studies have shown this statistical link and as the bacteria is very common and may just be a coinfectant agent of the flu.
The main hypothesis to explain the association between narcolepsy and influenza viruses is that narcolepsy is an autoimmune disease. Proteins of H1N1 virus may share homology with hypocretin neurons-overexpressed ones, bind to HLA-DQB1*06:02 and lead to the activation of autoreactive immune cells against hypocretin neurons. Even if these cells have not been yet identified, narcolepsy has been linked for the last decades with genes involved in the immune response
In this review, we will focus on the genes involved in the immune response and associated with narcolepsy. Most of these genes are implicated in the T-cell response. In a first part, we will present genes expressed by antigen-presenting cells (APCs) and involved in the processing and presentation of peptides to the T-cells. Then, we will focus on the T-cell receptor (TCR) which has been wells studied in the past years. Finally, genes associated with T cell survival and T cell immunosuppressive activity will be presented.
Narcolepsy is associated with polymorphism of genes implicated in antigen presentation by APC
Two proteins, cathepsin H and HLA-DQB1, are involved in antigen processing and presentation, and are genetically associated with narcolepsy (Figure 1) [13,14]••. In 2013, six single nucleotide polymorphims (SNPs) were discovered in cathepsin H (two in intron 1, a region already known to be associated with type 1 diabetes, and four in exon 1) by an immunochip study [13]••[15–17]. This analysis, performed on chips containing 200000 SNPs associated with nine auto-immune diseases, included 1886 narcoleptic patients and 10241 controls of European ancestry [13]••. One of the SNPs localized in exon 1, rs2289702T, is associated with a mutation (Gly11Arg) in the peptide signal of cathepsin H [13]••. Cathepsin H is a lysosomal cysteine proteinase activated by low pH, cleavage, and dimerization which acts as an endopeptidase or as an aminopeptidase [18]. This enzyme plays an important role in degradation of endocytosed proteins, cellular recycling, antigen processing and peptides loading onto HLA class II proteins [18]. Indeed, the absence of cathepsin H impairs immune cells development whereas a cathepsin H overexpression is observed during inflammation [19,20]. The activity of cathepsin H is extremely regulated by its natural inhibitors, as cystatin [13]••. The activity and the concentration of cathepsine H are partially dysregulated by Gly11Arg. This mutation introduces a polar amino acid positively charged in the peptide signal of cathepsin H. It modifies the trafficking and potentially the protein cleavage (bioinformatic simulations) leading to a decreased concentration of cathepsin H in the lysosomes [13]••. Therefore there may not be enough cathepsin H to correctly process the proteins impacting the protein repertory presented by HLA molecules.
Figure 1. Genes associated with narcolepsy encode proteins involved in antigen presentation (HLA class II pathway) and CD4+ T-cell activation.
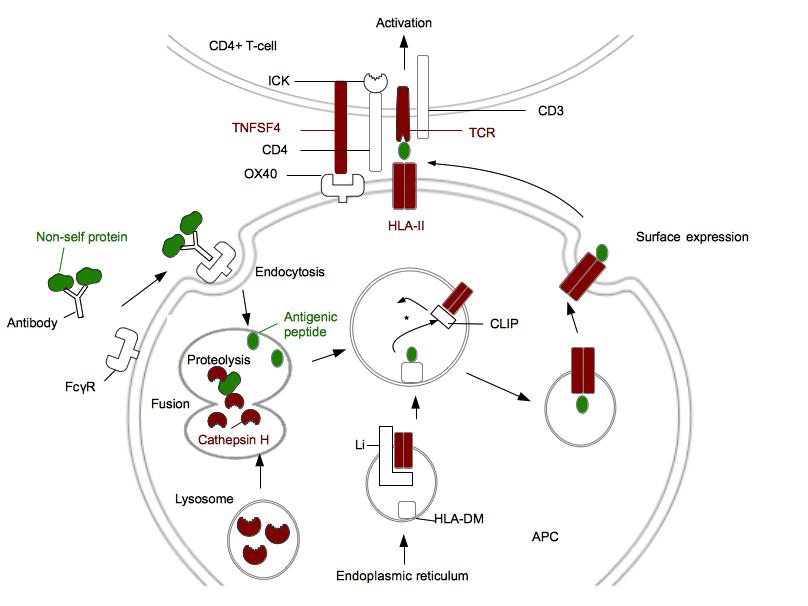
Proteins genetically associated with narcolepsy are in red. Non-self protein and antigenic peptide are in green. FcγR is a receptor for opsonized particles in APCs. HLA-DM are HLA-II like proteins required to facilitate the loading of the antigenic peptide [42]. *: In the HLA II compartment, the decrease of pH leads to the Li degradation in CLIP. CLIP will be replaced by the antigenic peptide. ICK is a kinase associated with CD4 which phosphorylates CD3 and leads to the T-cell activation. This figure does not represent P2RY11 and proteins involved in immunosuppressive activity of regulatory T-cells. (Adapted from [ 42])
Narcolepsy is also associated to HLA-DQB1*06:02 (Figure 2) [14]••. Interestingly, this allele is protective against type 1 diabetes meaning that for two different immune system-associated disease the same allele can have opposite effects [21]. HLA-DQB1 belongs to the HLA class II family and its product forms a heterodimer with HLA-DQA1, the product of the next door HLA gene on chromosome 6. The association of narcolepsy with this allele was found after that 509 narcoleptic patients with or without cataplexy were HLA typed [14]. More than 90% of patients with cataplexy were positive for HLA-DQB1*06:02 against 25% in ethnical matched controls [22]. For patients with mild, atypical or no cataplexy, the frequency of the allele was around 50%. The protein encoded by HLA-DQB1*0602 has nine polymorphic aminoacids in comparison to the protein encoded by HLA-DQB1*0601 , a protective allele for narcolepsy [21]. Five polymorphic amino acids are found in the binding pockets, keys for the HLA peptide interaction, and one of them is associated with the mutation Asp37Tyr. This mutation has an important impact on the HLA-peptide complex as Asp37Tyr is in pocket P9 and introduces a negative charge impacting the hydrogen bonding network [21]. Even if the polar character of P9 is conserved, this mutation seems to modify the binding specificity of P9 and to increase the affinity for the peptide. Furthermore, two other polymorphims found in the binding pocket P4 are associated with two mutations, Ala13Gly and Tyr26Leu [22]. These mutations decrease steric clashes within P4 and the interaction between P4 and the peptide is stabilized. These polymorphisms found in regions in contact with the peptide lead to an augmentation of the stability of the complex and an increase of the peptide presentation time at the membrane (Figure 1).
Figure 2. Narcolepsy is associated with HLA-DQB1*06:02 .
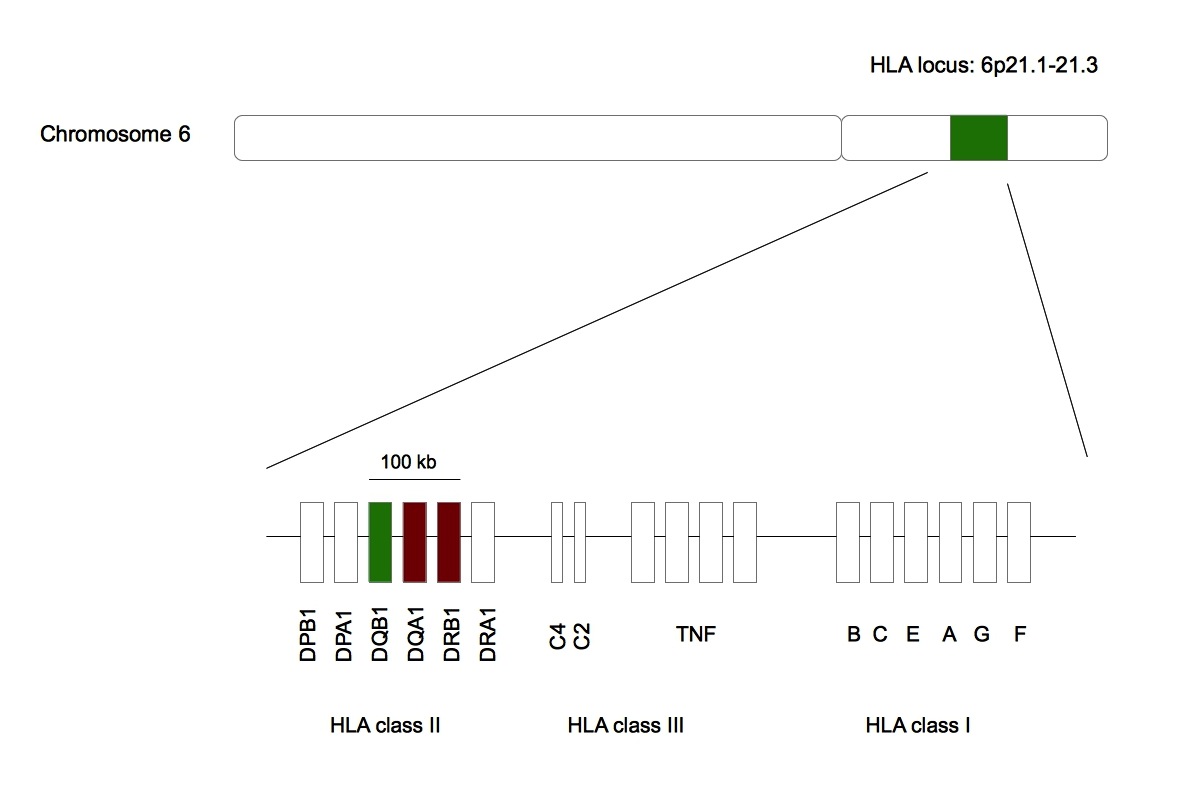
Green represents HLA-DQB1 whose allele is statistically linked with narcolepsy. Red represents HLA-DQA1 and HLA-DRB1 whose alleles are artificially associated with the sleep disorder. All these genes are found in the same 100 kb region in the HLA locus belonging to the chromosome 6 [43]•.
The statistical association between narcolepsy and HLA-DQB1*06:02 is so strong for patients with clear cataplexy that narcolepsy was initially linked in 1984 with an allele of another HLA gene very closed to HLA-DQB1 : HLA-DRB1*1501 [23]•. All the Japanese patients studied in this analysis were positive for this allele. Nevertheless, the link between narcolepsy and HLA-DRB1*1501 was less pronounced in other ethnic groups and was not found in African-American narcoleptic patients. Narcolepsy is also artificially linked with HLA-DQA1*01:02 as its products dimerizes with HLA-DQB1*06:02 [3]•.
TCR is genetically linked with narcolepsy
In the previous part, we showed that narcolepsy is strongly linked with HLA-DQB1*06:02. Nevertheless, Mignot et al. demonstrated in 1998 that the disease was not fully explained by polymorphisms found in the HLA locus as familial cases of narcolepsy may more often be HLA-DQB1*06:02 negative than sporadic cases: up to a third of narcoleptic families does not have this allele [24]. In order to know if other genes could explain the susceptibility to narcolepsy, genome-wide association study was performed in Caucasians from US, Canada and Europe centers for narcolepsy [25]••. Cases were all HLA-DQB1*06:02 positive with clear-cut narcolepsy-cataplexy and were geographically and ethnically matched with HLA-DQB1*06:02 positive controls. Cerebrospinal fluid hypocretin levels were available for 23% of narcoleptic cases and they were systematically insufficient in comparison to standard levels. In addition to these expected results, three markers in TCRα were found to be in high link disequilibrium with caucasian narcolepsy (Figure 3) [25]••. To confirm this discovery, the genome-wide association study was done again in two other ethnic groups (Asians and African-Americans): the same three markers were linked with narcolepsy and confirmed by further studies [26]•. TCRα encodes for the α chain of the TCRαβ heterodimer interacting with HLA class I and HLA class II molecules [27]. TCRα is made of variable, junction and constant regions and undergoes a random somatic recombination. Even if the molecular mechanisms of the link between narcolepsy and TCRα remain unknown, it is likely that the mutations associated with the SNPs have an effect on the somatic recombination as these SNPs are found in the junction region which is important for the recombination of the variable region with the constant region. The idea is that the recombination is not random in the presence of these SNPs leading to the formation of a TCRα very affine for the HLA-DQA1*01:02/HLA-DQB1*0602 peptide complex [13]••[25]••[26]•. Therefore, the activation of CD4+ T cells will be stronger and could lead to the destruction of hypocretin neurons if the non self presented peptide shares homology with proteins overexpressed in these neurons.
Figure 3. Localization of SNPs associated with narcolepsy in TCRα.
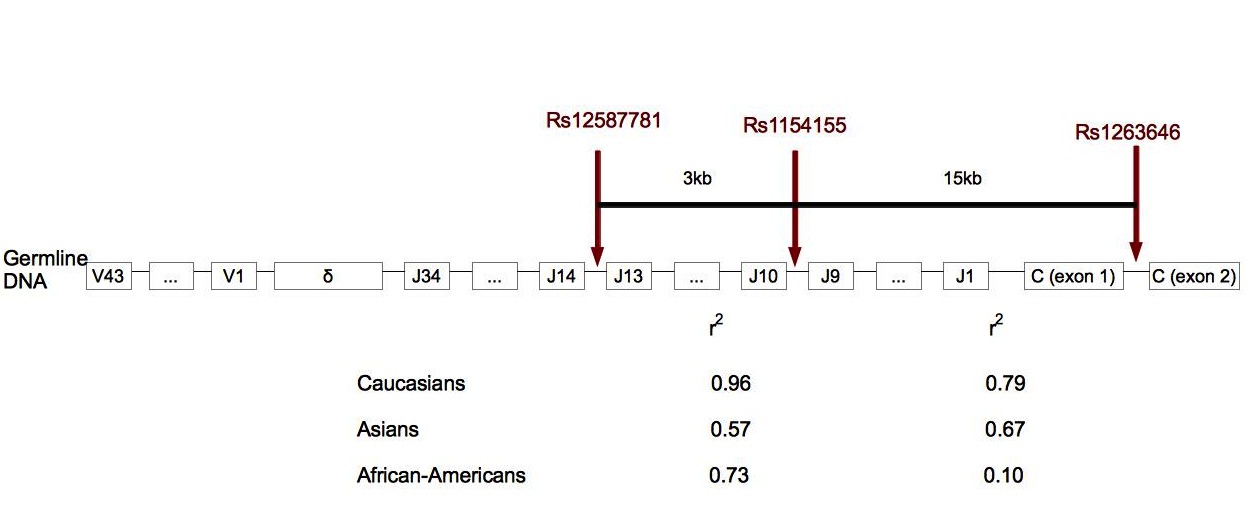
In human cells, TCRα locus is made of 43 variable regions, 34 junction regions and one constant region. TCRδ is localized in TCRα locus, between the variable and junction regions. Rs12567781 is localized between junction regions 13 and 14, rs1154155 between junction regions 9 and 10, and rs1263646 between exons 1 and 2 of the constant region. r2 gives the link disequilibrium between SNPs for each ethnic group. In Caucasians and African-Americans, the link between rs12587781 and rs1154155 is stronger than the link between rs1154155 and rs1263646. This suggests that the association peak is in the junction region rather than in the constant region. Rs1154155 is the marker the most significantly associated with narcolepsy [25]••. (Adapted from [25]).
Recently, an additional study on the association between narcolepsy and TCR genes showed that TCRβ was also involved [28]. 1189 narcoleptic patients and 1997 controls from China were included in the study. Narcoleptic cases were all diagnosed at the clinical center of Beijing University with hypocretin deficiency, clear-cut cataplexy and HLA-DQB1*06:02 positivity [28]. Three markers were discovered in the variable region of TCRβ . TCRβ undergoes a VDJ somatic recombination [29]. The presence of D segments increases the diversity of the recombinant TCRβ repertory in comparison to the TCRα repertory. The effects of the mutations associated with these SNPs are poorly known and understood. As previously, they may have an effect on the somatic recombination leading to the formation of a TCRβ affine for the HLA-DQA1*01:02/HLA-DQB1*06:02 peptide complex. The fact that these statistical associations with TCRα and TCRβ loci found in narcolepsy but not in other HLA-associated diseases such as type 1 diabetes may reflect a relative oligoclonality of the TCR subtypes involved in the physiopathology of narcolepsy, themselves tightly associated with a single HLA heterodimer.
Narcolepsy is associated with genes involved in the survival and the proliferation of T-cells and the immunosuppressive effect of regulatory T-cells.
After the study performed to find the association between TCR genes and narcolepsy, another genome-wide association study was done in a European ancestry population (594 narcoleptic cases against 931 controls) on 10 loci including genes involved in immune response and 18 markers, already known to be linked with narcolepsy or unknown, were found [30]. The highest significant SNP, rs4804122, was localized in the downstream region of P2RY11 . Interestingly the same study was replicated in two other ethnic groups (Asians and African-Americans), but rs4804122 was not found to be associated with narcolepsy [30]. Indeed, it was another SNP from the initial 18 markers, rs2305795 localized in the 3' UTR of P2RY11 , which showed the highest association with narcolepsy across all ethnic groups [30]. Rs2305795 is associated with one or two contiguous mutations in the 3' UTR of P2RY11 : GG becomes AG or AA. This polymorphism had also been found in Chinese narcolepsy [26]•. P2RY11 is a low affinity receptor for ATP belonging to a family of more than 20 purinergic receptors [31]. The expression of P2RY11 is widespread in all immune cells and its function is unusual: P2RY11 activation counterbalances the cytotoxic effect of ATP mediated by other purinergic receptors [30–32]. In CD8+ T-cells, P2RY11 expression is higher than in other immune cells and it also protects against cytotoxic effects of ATP [30]. The mutation associated with rs2305795 decreases this protection and leads to a decreased CD8+ T-cells survival [30]. Seemingly, this result is surprising because autoimmunity is a process involving proliferation of immune cells and because the link between narcolepsy and CD8+ T-cells is not clear. Therefore, the discovery of these SNP suggests other P2RY11 functions presently unknown [30]. The link between narcolepsy and CD4+ T-cells is more clear because CD4+ T-cells express a TCR recognizing HLA class II molecules such as HLA-DQB1*06:02. Interestingly, polymorphisms have been found in genes involved in CD4+ T-cells survival and proliferation.
The immunochip study previously described led to the discovery of another SNP, rs7553711, strongly associated with narcolepsy and located in the enhancer of the Tumor Necrosis Factor Superfamily Member 4 ( TNFSF4 ) [13]. Other SNPs were found in TNFSF4 but were weakly associated with narcolepsy. Interestingly, these latest SNPs are strongly associated with other autoimmune diseases, as systemic lupus erythematosis or systemic sclerosis [33–35]. TNFSF4 is a membranous protein expressed by CD4+ T-cells interacting with its ligand on APC plasmic membranes (Figure 1) [3,13]••. The interaction between TNFSF4 and its ligand is a coactivation signal for T-cells: it leads to the survival and proliferation of effector T-cells and generation of memory T-cells [13]••. Excessive quantity of TNFSF4 leads to a disruption of tolerance [36]. The molecular mechanisms of the link between destruction of hypocretin neurons and TNFSF4 are unknown. The actual hypothesis is that the polymorphism associated with rs7553711 is likely involved in the over expression of TNFSF4 because of the localization of rz7553711 in the enhancer region. This over expression of TNFSF4 would promote the survival and the proliferation of CD4+ T-cells [13]••. Therefore, TNFS4 will indirectly activate the immune response. Proliferation of T-cells is extremely regulated by a sub-population of CD4+ T-cells, regulatory T-cells. These regulatory T-cells have an immunosuppressive effect preventing autoimmune processes. Two genes expressed by regulatory T-cells were found to be linked with narcolepsy.
The first one is DNMT1 and was discovered in five narcoleptic patients with other disorders. This set of disorders is called autosomal dominant cerebellar ataxia, deafness and narcolepsy (ADCA-DN) [37]••. This form of narcolepsy is different from the sporadic one, is rarer and appears in the adulthood. Exome sequencing led to the identification of three different mutations all found in the exon 21 of DNMT1 (Figure 4A) [37]••. Sanger sequencing of narcoleptic patients confirmed the presence of these mutations in DNMT1 . DNMT1 is a DNA methyltransferase involved in gene silencing through CpG methylation and direct binding of HDAC2 [38]. This enzyme is involved in the differentiation of CD4+ T-cells into regulatory T-cells [15]. The three mutations affect regions involved in the interaction between DNMT1 and DNA or between DNMT1 and HDAC2 (Figure 4B). Therefore, these mutations could indirectly affect the differentiation of CD4+ T-cells into regulatory T-cells.
Figure 4. Localization of the three mutations in DNMT1 sequence (A) and their involvement in DNMT1-DNA interaction
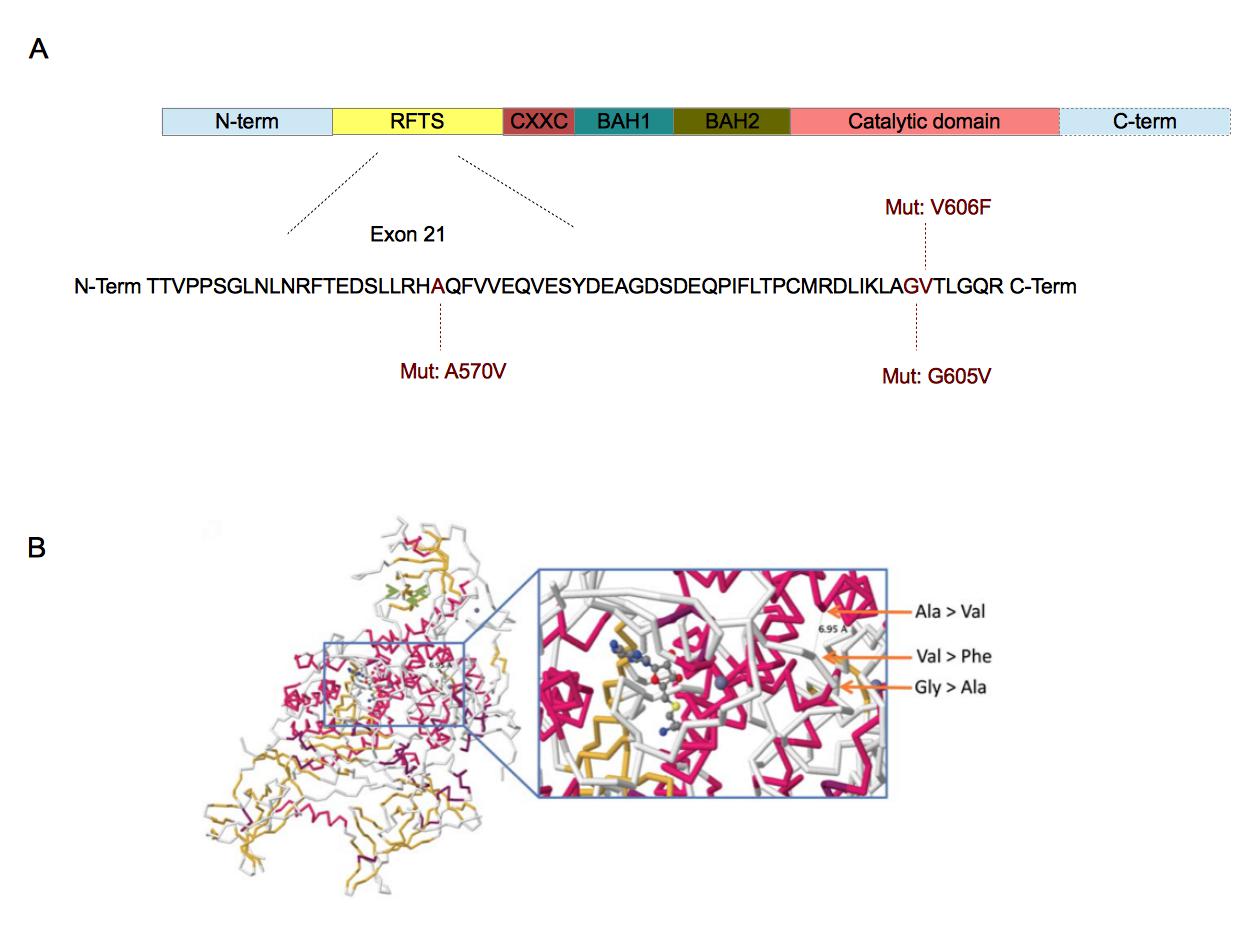
A. DNMT1 is composed of five domains. The replication focus targeting sequence (RFTS) domain is a DNA-competitive inhibitor for DNMT1 [44]. CXXC domain recognizes CpG sequences [38]. BAH1 and BAH2 domains are involved in protein-protein interactions [38]. The catalytic domain is associated with the methyltransferase activity [37]••. The three mutations have been found in the exon 21 which is part of the RFTS domain [37]••. The amino acids involved are well conserved across vertebrates [37]••. B. These mutations are closed from each other in the 3D structure of DNMT1. Furthermore, they are also closed from the catalytic domain of DNMT1. The substrate showed in this crystallographic reconstruction is S-adenosyl-L-homocystein. ( B. Adapted from [37]••).
The second gene associated with the immunosuppressive activity of regulatory T-cells and genetically involved in narcolepsy is Interleukine 10 Receptor β (IL10Rβ) . Two novel SNPs, rs2834190 and rs2409488, were found in the region of IL10RB [28]. This genetic association between narcolepsy and IL10Rβ was only found in one study therefore the involvement of IL10Rβ is still hypothetical. IL10Rβ is the β chain of the IL10 receptor, IL10 being an immunosuppressive cytokine synthesized by regulatory T-cells. Interestingly, IL10Rβ is genetically associated with Crohn disease and involved in inflammatory bowel disease [39,40]. The mutation associated with these SNPs, would probably alter the interaction between IL10 and IL10R. Nevertheless, the association between IL10Rβ and narcolepsy has not yet been confirmed and there is no additional data on this SNP to confirm the theoretical model.
Conclusion
The loss of hypocretin neurons in narcolepsy results from a sequence of events. The overarching susceptibility stems from the HLA-DQB1*06:02/ HLA-DQA1*01:02 heterodimer, as nearly all cases are HLA-DQB1*06:02 positive. Additional susceptibility is associated with variant in genes involved in the immune response such as T cell receptor alpha , T cell receptor β and cathepsin H (Table 1). Nevertheless, narcolepsy is not a purely Mendelian disorder as only 32% of monozygotic twins are concordant for this disorder (6 out 19 twin pairs reported for this disease) [41]. In this context, environmental factors as influenza A and Streptococcus pyogenes infections or H1N1 vaccination are keys and could spur an autoimmune process leading to the destruction of hypocretin neurons.
Nevertheless, the autoreactive immune cells and the molecular mimicry between hypocretin neurons overexpressed proteins and H1N1 proteins are still unknown. A key next step could be to use CD4+ T-cells from patients and controls and to test their reactivity to H1N1 peptides by enzyme-linked immunospot (ELISpot). Then, viral peptides specifically activating narcoleptic patients CD4+ T-cells could be in silico aligned against hypocretin neurons-overexpressed proteins in order to identify a possible mimicry. As narcolepsy development was more associated with 2009 H1N1 pandemic than with seasonal flu strains, it could be interesting to verify if these viral peptides are 2009 specific. Finally, these viral epitopes should also be aligned against Streptococcus pyogenes proteins as the link between streptococcal infections and narcolepsy remains controversial.
Table 1. Summary of major polymorphisms in genes involved in immune response and associated with narcolepsy.
|
Gene |
Localization of the gene |
Type and name of genetic variant |
Localization of the variant in the gene genetic |
Theoretical effect of variant |
|
Cathepsin H [13] |
15q25.1 |
SNP: rs2289702 (Gly11Arg) |
Region encoding for the peptide signal |
Alteration of the protein repertory presented by APC |
|
6p21.1-21.3 [14] |
Allele: HLA-DQB1*0602 (Ala13Gly, Tyr26Leu, Asp37Tyr)[21] |
Regions encoding for pockets P4 and P9 [21] |
Increasing stability of HLA-hypocretin complexes [21] |
|
|
TCRα [25] |
14q11.2 |
SNP: Rs12587781 (NC1) Rs1154155 (NC) Rs1263646 (NC) |
Junction region and constant region |
Recombination of an affine TCRα for HLA-DQB1*06:02/HLA-DQA1*01:02 peptide complex |
|
TCRβ. [28] |
7q34 |
SNP: Rs9648789 (NC) Rs9648789 (NC) Rs3020837 (NC) |
Variable region |
Recombination of an affine TCRβ for HLA-DQB1*06:02/HLA-DQA1*01:02 peptide complex |
|
P2RY11 [13] |
19p13.2 |
SNP: Rs2305795 (NC) |
3' UTR |
Decreased survival of CD8+ T-cells, other effect? |
|
TNFSF4 [13] |
1q25.1 |
SNP: Rs7553711 (NC) |
Enhancer |
Over survival / proliferation of CD4+ T-cells |
|
DNMT1 [37] |
19p13.2 |
Mutation: A570, G605V, V606F |
Region encoding for the RFTS domain |
Alteration of DNMT1-DNA interaction |
|
IL10Rβ [28] |
21q22.11 |
SNP: Rs2409488 Rs2834190 |
5' region of the gene |
Alteration of IL10-IL10R interaction |
1: NC (non coding) indicates that the SNP is in a non coding region.
Acknowledgments
I thank Professor Emmanuel Mignot and Dr Nathalie Alazard-Dany for their help and their advises concerning this review.
References and recommended reading
Papers of particular interest have been highlighted as:
● of special interest
●● of outstanding interest
1. Overeem S, Black JL, Lammers GJ: Narcolepsy: immunological aspects . Sleep Med. Rev. 2008, 12 :95–107. [ Full Paper ][ PubMed ][ PubMed Central ]
2. Okun ML, Lin L, Pelin Z, Hong S, Mignot E: Clinical aspects of narcolepsy-cataplexy across ethnic groups . Sleep 2002, 25 :27–35. [ PubMed ]
3.
Mahlios J, De la Herrán-Arita AK, Mignot E:
The autoimmune basis of narcolepsy
.
Curr. Opin.
Neurobiol.
2013,
23
:767–773. [
Full Paper
]
● This review summarized genetic,
immunological and environmental factors associated with narcolepsy. The
functional effects of polymorphisms in genes involved in the immune response
were not developed in this paper.
4. Pizza F, Vandi S, Detto S, Poli F, Franceschini C, Montagna P, Plazzi G: Different sleep onset criteria at the multiple sleep latency test (MSLT): an additional marker to differentiate central nervous system (CNS) hypersomnias . J. Sleep Res. 2011, 20 :250–256. [ Full Paper ]
5. Pizza F, Franceschini C, Peltola H, Vandi S, Finotti E, Ingravallo F, Nobili L, Bruni O, Lin L, Edwards MJ, et al.: Clinical and polysomnographic course of childhood narcolepsy with cataplexy . Brain J. Neurol. 2013, 136 :3787–3795. [ Full Paper ][ PubMed ]
6.
Chemelli RM, Willie JT, Sinton CM, Elmquist JK, Scammell T, Lee C,
Richardson JA, Williams SC, Xiong Y, Kisanuki Y, et al.:
Narcolepsy in orexin knockout mice: molecular genetics of
sleep regulation
.
Cell
1999,
98
:437–451. [
PubMed
]
●● In this paper, authors showed
that a hypocretin knockout mice had a narcolepsy-like phenotype. It was the
first paper showing that narcolepsy may be caused by a loss of hypocretin.
7. Tsujino N, Sakurai T: Orexin/Hypocretin: A Neuropeptide at the Interface of Sleep, Energy Homeostasis, and Reward System . Pharmacol. Rev. 2009, 61 :162–176. [ Full Paper ][ PubMed ]
8. Tsujino N, Sakurai T: Role of orexin in modulating arousal, feeding, and motivation [Internet] . Front. Behav. Neurosci. 2013, 7 . [ Full Paper ][ PubMed ][ PubMed Central ]
9. Heier MS, Jansson TS, Gautvik KM: Cerebrospinal fluid hypocretin 1 deficiency, overweight, and metabolic dysregulation in patients with narcolepsy . J. Clin. Sleep Med. JCSM Off. Publ. Am. Acad. Sleep Med. 2011, 7 :653–658. [ Full Paper ][ PubMed ]
10. Andlauer O, Moore H, Hong S-C, Dauvilliers Y, Kanbayashi T, Nishino S, Han F, Silber MH, Rico T, Einen M, et al.: Predictors of Hypocretin (Orexin) Deficiency in Narcolepsy Without Cataplexy . Sleep 2012, 35 :1247–1255. [ Full Paper ][ PubMed ][ PubMed Central ]
11. Han
F, Lin L, Warby SC, Faraco J, Li J, Dong SX, An P, Zhao L, Wang LH, Li QY, et
al.:
Narcolepsy onset is seasonal
and increased following the 2009 H1N1 pandemic in china
.
Ann.
Neurol.
2011,
70
:410–417. [
Full Paper
]
●● This Chinese study showed that
the incidence of narcolepsy increased few months after the beginning of 2009
pandemic. The incidence increase was higher than the one observed after
seasonal flu.
12.
Aran A, Lin L, Nevsimalova S, Plazzi G, Hong SC, Weiner K, Zeitzer J,
Mignot E:
Elevated
anti-streptococcal antibodies in patients with recent narcolepsy
onset
.
Sleep
2009,
32
:979–983. [
PubMed
][
PubMed Central
]
● This paper remains controversial since the
association between narcolepsy and Streptococcus pyogenes infections was not
confirmed by other studies. It is possible that Streptococcus pyogenes is a
coinfectant of influenza viruses and that the link found by the author is
artifactual.
13. Faraco J, Lin L, Kornum BR,
Kenny EE, Trynka G, Einen M, Rico TJ, Lichtner P, Dauvilliers Y, Arnulf I, et al.:
ImmunoChip study implicates antigen presentation to t cells in
narcolepsy [Internet]
.
PLoS Genet.
2013,
9
. [
Full
Paper
][
PubMed
][
PubMed Central
]
●● This genetic study was important for the identification of
narcolepsy-associated genes other than
HLA-DQB1
14.
Mignot E, Hayduk R, Black J, Grumet FC, Guilleminault C:
HLA DQB1*0602 is associated with
cataplexy in 509 narcoleptic patients
.
Sleep
1997,
20
:1012–1020.
[
PubMed
]
●● Mignot showed in this paper that narcolepsy was linked with
HLA-DQB1
and not with
HLA-DRB1
.
15. Singh AK, Mahlios J, Mignot E: Genetic association, seasonal infections and autoimmune basis of narcolepsy . J. Autoimmun. 2013, 43 :26–31. [ Full Paper ]
16. Cooper JD, Smyth DJ, Smiles AM, Plagnol V, Walker NM, Allen JE, Downes K, Barrett JC, Healy BC, Mychaleckyj JC, et al.: Meta-analysis of genome-wide association study data identifies additional type 1 diabetes risk loci . Nat. Genet. 2008, 40 :1399–1401. [ Full Paper ][ PubMed ][ PubMed Central ]
17. Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA, Julier C, Morahan G, Nerup J, Nierras C, et al.: Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes . Nat. Genet. 2009, 41 :703–707. [ Full Paper ] [PMID: 19430480] [ PubMed Central ]
18. Conus S, Simon H: Cathepsins and their involvement in immune responses [Internet] . Swiss Med. Wkly. 2010, doi:10.4414/smw.2010.13042. [ Full Paper ]
19. Colbert JD, Matthews SP, Miller G, Watts C: Diverse regulatory roles for lysosomal proteases in the immune response . Eur. J. Immunol. 2009, 39 :2955–2965. [ Full Paper ][ PubMed ]
20. Li X, Wu K, Edman M, Schenke-Layland K, MacVeigh-Aloni M, Janga SR, Schulz B, Hamm-Alvarez SF: Increased expression of cathepsins and obesity-induced proinflammatory cytokines in lacrimal glands of male NOD mouse . Invest. Ophthalmol. Vis. Sci. 2010, 51 :5019–5029. [ Full Paper ][ PubMed ][ PubMed Central ]
21. Siebold C, Hansen BE, Wyer JR, Harlos K, Esnouf RE, Svejgaard A, Bell JI, Strominger JL, Jones EY, Fugger L: Crystal structure of HLA-DQ0602 that protects against type 1 diabetes and confers strong susceptibility to narcolepsy . Proc. Natl. Acad. Sci. U. S. A. 2004, 101 :1999–2004. [ Full Paper ][ PubMed ]
22. Mignot E, Young T, Lin L, Finn L: Nocturnal sleep and daytime sleepiness in normal subjects with HLA-DQB1*0602 . Sleep 1999, 22 :347–352. [ PubMed ]
23.
Juji T, Satake M, Honda Y, Doi Y:
HLA antigens in Japanese patients with narcolepsy. All the
patients were DR2 positive
.
Tissue
Antigens
1984,
24
:316–319. [
PubMed
]
●
HLA-DRB1
was presented to
be associated with narcolepsy in this study. This effect is an artifact.
That is explained by the proximity of
HLA-DR*B1
with
HLA-DQ*B1
24. Mignot E: Genetic and familial aspects of narcolepsy . Neurology 1998, 50 :S16–22. [ PubMed ]
25.
Hallmayer J, Faraco J, Lin L, Hesselson S, Winkelmann J, Kawashima M,
Mayer G, Plazzi G, Nevsimalova S, Bourgin P, et al.:
Narcolepsy is strongly associated with the TCR alpha
locus
.
Nat. Genet.
2009,
41
:708–711. [
Full Paper
][
PubMed
][
PubMed Central
]
●● Hallmayer and al showed that narcolepsy is associated with the
T cell receptor
alpha
.
26. Han
F, Lin L, Li J, Aran A, Dong SX, An P, Zhao L, Li QY, Yan H, Wang JS, et al.:
TCRA, P2RY11, and CPT1B/CHKB
associations in Chinese narcolepsy
.
Sleep Med.
2012,
13
:269–272. [
Full Paper
][
PubMed
][
PubMed Central
]
● Another study of the genes
implicated in the immune response and associated with the sleep
disorder.
27. Huang J, Meyer C, Zhu C: T cell antigen recognition at the cell membrane . Mol. Immunol. 2012, 52 :155–164. [ Full Paper ][ PubMed ][ PubMed Central ]
28. Han F, Faraco J, Dong XS, Ollila HM, Lin L, Li J, An P, Wang S, Jiang KW, Gao ZC, et al.: Genome wide analysis of narcolepsy in China implicates novel immune loci and reveals changes in association prior to versus after the 2009 H1N1 Influenza pandemic [Internet] . PLoS Genet. 2013, 9 . [ Full Paper ][ PubMed ][ PubMed Central ]
29. Bonnet M, Ferrier P, Spicuglia S: Molecular genetics at the T-cell receptor beta locus: insights into the regulation of V(D)J recombination . Adv. Exp. Med. Biol. 2009, 650 :116–132. [ PubMed ]
30. Kornum BR, Kawashima M, Faraco J, Lin L, Rico T, Hesselson S, Axtell RC, Kuipers H, Weiner K, Hamacher A, et al.: Common variants in P2RY11 are associated with narcolepsy . Nat. Genet. 2011, 43 :66–71. [ Full Paper ][ PubMed ][ PubMed Central ]
31. Van Der Weyden L, Conigrave AD, Morris MB: Signal transduction and white cell maturation via extracellular ATP and the P2Y11 receptor . Immunol. Cell Biol. 2000, 78 :369–374. [ Full Paper ]
32. Moore DJ, Chambers JK, Wahlin JP, Tan KB, Moore GB, Jenkins O, Emson PC, Murdock PR: Expression pattern of human P2Y receptor subtypes: a quantitative reverse transcription-polymerase chain reaction study . Biochim. Biophys. Acta 2001, 1521 :107–119. [ PubMed ]
33. Bossini-Castillo L, Broen JCA, Simeon CP, Beretta L, Vonk MC, Ortego-Centeno N, Espinosa G, Carreira P, Camps MT, Navarrete N, et al.: A replication study confirms the association of TNFSF4 (OX40L) polymorphisms with systemic sclerosis in a large European cohort . Ann. Rheum. Dis. 2011, 70 :638–641. [ Full Paper ][ PubMed ]
34. Cunninghame Graham DS, Graham RR, Manku H, Wong AK, Whittaker JC, Gaffney PM, Moser KL, Rioux JD, Altshuler D, Behrens TW, et al.: Polymorphism at the TNF superfamily gene TNFSF4 confers susceptibility to systemic lupus erythematosus . Nat. Genet. 2008, 40 :83–89. [ Full Paper ][ PubMed ][ PubMed Central ]
35. Gourh P, Arnett FC, Tan FK, Assassi S, Divecha D, Paz G, McNearney T, Draeger H, Reveille JD, Mayes MD, et al.: Association of TNFSF4 (OX40L) polymorphisms with susceptibility to systemic sclerosis . Ann. Rheum. Dis. 2010, 69 :550–555. [ Full Paper ][ PubMed ][ PubMed Central ]
36. Ishii N, Takahashi T, Soroosh P, Sugamura K: OX40-OX40 ligand interaction in T-cell-mediated immunity and immunopathology . Adv. Immunol. 2010, 105 :63–98. [ Full Paper ][ PubMed ]
37.
Winkelmann J, Lin L, Schormair B, Kornum BR, Faraco J, Plazzi G,
Melberg A, Cornelio F, Urban AE, Pizza F, et al.:
Mutations in DNMT1 cause autosomal dominant cerebellar
ataxia, deafness and narcolepsy
.
Hum. Mol.
Genet.
2012,
21
:2205–2210.
[
Full Paper
][
PubMed
][
PubMed Central
]
●● This paper is very interesting
because it presents a rarer form of narcolepsy found in a polysyndromic
disease. This autosomal dominant form of narcolepsy is associated with DNMT1
mutations probably impairing the regulatory T cells development.
38. Svedružić ŽM: Dnmt1 structure and function . Prog. Mol. Biol. Transl. Sci. 2011, 101 :221–254. [ Full Paper ][ PubMed ]
39. Mao H, Yang W, Lee PPW, Ho MH-K, Yang J, Zeng S, Chong C-Y, Lee T-L, Tu W, Lau Y-L: Exome sequencing identifies novel compound heterozygous mutations of IL-10 receptor 1 in neonatal-onset Crohn's disease . Genes Immun. 2012, 13 :437–442. [ Full Paper ][ PubMed ]
40. Begue B, Verdier J, Rieux-Laucat F, Goulet O, Morali A, Canioni D, Hugot J-P, Daussy C, Verkarre V, Pigneur B, et al.: Defective IL10 signaling defining a subgroup of patients with inflammatory bowel disease . Am. J. Gastroenterol. 2011, 106 :1544–1555. [ Full Paper ][ PubMed ]
41. Dauvilliers Y, Arnulf I, Mignot E: Narcolepsy with cataplexy . Lancet 2007, 369 :499–511. [ Full Paper ][ PubMed ]
42. Male DK: Immunology: An Illustrated Outline . Elsevier Health Sciences; 2004.1. Overeem S, Black JL, Lammers GJ: Narcolepsy: immunological aspects . Sleep Med. Rev. 2008, 12 :95–107. [ Full Paper ][ PubMed ][ PubMed Central ]
43.
Mignot E, Lin X, Arrigoni J, Macaubas C, Olive F, Hallmayer J,
Underhill P, Guilleminault C, Dement WC, Grumet FC:
DQB1*0602 and DQA1*0102 (DQ1) are better markers than DR2
for narcolepsy in Caucasian and black Americans
.
Sleep
1994,
17
:S60–67.
[
PubMed
]
● This paper was a confirmation of the other Mignot's study. Whereas
HLA-DRB1
is not linked
with narcolepsy in African-Americans,
HLA-DQB1
is well associated with the sleep disorder all
across ethnic groups.
44.Syeda F, Fagan RL, Wean M, Avvakumov GV, Walker JR, Xue S, Dhe-Paganon S, Brenner C: The replication focus targeting sequence (RFTS) domain is a DNA-competitive inhibitor of Dnmt1 . J. Biol. Chem. 2011, 286 :15344–15351. [ Full Paper ][ PubMed ][ PubMed Central ]



